Gene
KWMTBOMO12385 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014083
Annotation
PREDICTED:_metaxin-1-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.43
Sequence
CDS
ATGGCTAACATTGAATTAGACATTTGGAGAGGAGAATGGGGTCTAGCATCTATCGATTTAGAATGTTTAAAAGTATTAACTTACATGAAATTTATCGGTGTGCCTGTTAAAGTGAGAGAGTCGAATAACCCATTTTTCACCCCCAAAGGTCGACTTCCTGTCATGAGAGATGGACGGACCGTTCTTACAAATTTTGAAGAAGTAGTAGAACATTTGAAATCTCTACATTACAACACAGATGTACATTTAAACTCCAAAGAGGCAGCGGAAGCGAGTGCATTCACACAATACCTCCGAGACAAGTTATACCCAGCTTACCAGTATGCTTGGTGGGTGGATGAAAAGAATTACAATGATGTAACTAGGCCAACATTTGCAAAATGTTTGAGAATTCCATTTAATTTTTTTTATCCATCACGGTATCAGAATGCCGCCAAAGAAATGATAGACGCATTGTATGGAGAACATACTGATCTTAGAGAAATAGAGAAAACTATATACAGTGAAGCTGAGAAGTGCCTTAAAACATTATCAGATAGATTAGGTGAGAGTGAATACTTCTTTGGAAACCGTCCTTCATCGTTTGACGCTATTGTGTTTGCTTACTTAGCTCCTTTAGTGAAAGCACCATTTCCTAATGGGACTCTGTCAAACCATGTCAAAGGCACTGCAAATTTGGCAAGATTTGTGGCCAGGATCAATCAGAAAAATTTTAGACACGTCACAGATGAATATAATAAACAGAATGCAAAGAAACAGTCCACAGGCTCGCAGTCTGATAGAGAAGCCGCAAACTTTCCCAACCAGACTCGAAATAAAATCTTAGCAGCCCTGTTTGCTGCTATTGCTATGACGGGGTATGCAGTGGCTAATGGAATGTTTCAAGAGCTGAAAGACTTTGAGCACTCGCGAGATTACAATGAAATGTTTGAGAATGAAGAAGAGGACAACTGA
Protein
MANIELDIWRGEWGLASIDLECLKVLTYMKFIGVPVKVRESNNPFFTPKGRLPVMRDGRTVLTNFEEVVEHLKSLHYNTDVHLNSKEAAEASAFTQYLRDKLYPAYQYAWWVDEKNYNDVTRPTFAKCLRIPFNFFYPSRYQNAAKEMIDALYGEHTDLREIEKTIYSEAEKCLKTLSDRLGESEYFFGNRPSSFDAIVFAYLAPLVKAPFPNGTLSNHVKGTANLARFVARINQKNFRHVTDEYNKQNAKKQSTGSQSDREAANFPNQTRNKILAALFAAIAMTGYAVANGMFQELKDFEHSRDYNEMFENEEEDN
Summary
Uniprot
H9JX18
A0A3S2LUN0
A0A2H1WA37
A0A1E1WFS7
A0A2A4JIB5
A0A194Q110
+ More
A0A2W1BWX3 A0A212EZA5 A0A0L7LTA3 A0A194R9E4 A0A067RT21 A0A1B6E1Z4 A0A1B6CCM3 A0A2J7R506 A0A158NUL7 A0A195FKU3 A0A195BE71 A0A0P4X8H2 A0A0P5LYQ4 A0A0P6I9M2 A0A3L8DKH7 A0A151IFW2 A0A0J7KE41 E0VSG3 F4WRE9 A0A151JQ89 A0A088AFM0 A0A0N8BRY3 A0A0V0G761 V9IIZ0 A0A2A3EAU6 R4WIF9 A0A0L7R4T6 A0A1B6FSB1 A0A1B6EU28 A0A1B6KUB2 A0A1B6LGD4 A0A1B6KXI1 A0A0P4VJ10 R4G7S9 A0A0P5P8L6 A0A0P4ZUJ5 A0A0A9Z5R4 A0A232F340 K7J7T9 A0A2R7VY52 E2AFV4 A0A0M9A925 A0A087SZ12 W4YKT9 L7M3A2 A0A224YTA5 A0A131YL76 A0A224YT94 A0A131XFI5 A0A026W739 V3ZV28 B7Q5R0 A0A0K8RK28 A0A1Z5KY75 A0A0L8FS04 A0A293MYN1 A0A1E1XV46 A0A023GKJ7 G3MR45 A0A2C9LN35 A0A0C9S2E2 A0A443RBF0 A0A443SDA2 A0A3Q2DYX5 A0A147ASI4 A0A3S0ZDW7 K1PP76 A0A3Q2R0R9 A0A3Q1FVF1 H2MQS3 A0A0B6Y9N5 A0A2U9CIL0 A0A3P9CLF7 A0A1S3HHX9 A0A3Q1D8P2 I3J1F8 A0A146Y5N2 A0A3P8PLA0 A0A3B5ARS9 A0A1A8U7K9 A0A1A8F3H6 A0A3P8TWP4 A0A3P9KM60 A0A1A8IY73 A0A3P9JHG0 A0A1A8RKU8 A0A1A8NNE6 A0A1A8DKQ7 A0A3Q3MLB6 A0A3Q3GZT9 A0A1A7YD69 A0A3Q0S8X7 A0A3Q3K0Y7
A0A2W1BWX3 A0A212EZA5 A0A0L7LTA3 A0A194R9E4 A0A067RT21 A0A1B6E1Z4 A0A1B6CCM3 A0A2J7R506 A0A158NUL7 A0A195FKU3 A0A195BE71 A0A0P4X8H2 A0A0P5LYQ4 A0A0P6I9M2 A0A3L8DKH7 A0A151IFW2 A0A0J7KE41 E0VSG3 F4WRE9 A0A151JQ89 A0A088AFM0 A0A0N8BRY3 A0A0V0G761 V9IIZ0 A0A2A3EAU6 R4WIF9 A0A0L7R4T6 A0A1B6FSB1 A0A1B6EU28 A0A1B6KUB2 A0A1B6LGD4 A0A1B6KXI1 A0A0P4VJ10 R4G7S9 A0A0P5P8L6 A0A0P4ZUJ5 A0A0A9Z5R4 A0A232F340 K7J7T9 A0A2R7VY52 E2AFV4 A0A0M9A925 A0A087SZ12 W4YKT9 L7M3A2 A0A224YTA5 A0A131YL76 A0A224YT94 A0A131XFI5 A0A026W739 V3ZV28 B7Q5R0 A0A0K8RK28 A0A1Z5KY75 A0A0L8FS04 A0A293MYN1 A0A1E1XV46 A0A023GKJ7 G3MR45 A0A2C9LN35 A0A0C9S2E2 A0A443RBF0 A0A443SDA2 A0A3Q2DYX5 A0A147ASI4 A0A3S0ZDW7 K1PP76 A0A3Q2R0R9 A0A3Q1FVF1 H2MQS3 A0A0B6Y9N5 A0A2U9CIL0 A0A3P9CLF7 A0A1S3HHX9 A0A3Q1D8P2 I3J1F8 A0A146Y5N2 A0A3P8PLA0 A0A3B5ARS9 A0A1A8U7K9 A0A1A8F3H6 A0A3P8TWP4 A0A3P9KM60 A0A1A8IY73 A0A3P9JHG0 A0A1A8RKU8 A0A1A8NNE6 A0A1A8DKQ7 A0A3Q3MLB6 A0A3Q3GZT9 A0A1A7YD69 A0A3Q0S8X7 A0A3Q3K0Y7
Pubmed
EMBL
BABH01042711
RSAL01000188
RVE44754.1
ODYU01007280
SOQ49913.1
GDQN01005333
+ More
JAT85721.1 NWSH01001364 PCG71539.1 KQ459591 KPI97085.1 KZ149932 PZC77310.1 AGBW02011326 OWR46822.1 JTDY01000133 KOB78685.1 KQ460473 KPJ14463.1 KK852433 KDR23960.1 GEDC01026821 GEDC01005346 JAS10477.1 JAS31952.1 GEDC01026116 JAS11182.1 NEVH01007395 PNF35918.1 ADTU01026592 KQ981522 KYN40614.1 KQ976504 KYM82868.1 GDIP01246214 JAI77187.1 GDIQ01166441 JAK85284.1 GDIQ01007635 JAN87102.1 QOIP01000007 RLU20930.1 KQ977771 KYN00032.1 LBMM01008979 KMQ88489.1 DS235750 EEB16319.1 GL888285 EGI63220.1 KQ978660 KYN29395.1 GDIQ01150795 JAL00931.1 GECL01002319 JAP03805.1 JR047032 AEY60309.1 KZ288301 PBC28845.1 AK417190 BAN20405.1 KQ414657 KOC65786.1 GECZ01016689 JAS53080.1 GECZ01028274 JAS41495.1 GEBQ01024942 JAT15035.1 GEBQ01017289 JAT22688.1 GEBQ01023820 JAT16157.1 GDKW01002508 JAI54087.1 ACPB03021239 GAHY01002192 JAA75318.1 GDIQ01134104 JAL17622.1 GDIP01209451 JAJ13951.1 GBHO01004388 GBRD01012714 JAG39216.1 JAG53110.1 NNAY01001090 OXU25174.1 KK854153 PTY12259.1 GL439158 EFN67703.1 KQ435710 KOX79700.1 KK112616 KFM58101.1 AAGJ04113373 GACK01007436 JAA57598.1 GFPF01009691 MAA20837.1 GEDV01009272 JAP79285.1 GFPF01009692 MAA20838.1 GEFH01002317 JAP66264.1 KK107390 EZA51436.1 KB202719 ESO88232.1 ABJB010182666 ABJB011105958 ABJB011127826 DS863069 EEC14182.1 GADI01002358 JAA71450.1 GFJQ02007166 JAV99803.1 KQ427052 KOF67464.1 GFWV01020320 MAA45048.1 GFAA01000276 JAU03159.1 GBBM01001845 JAC33573.1 JO844346 AEO35963.1 GBZX01001581 JAG91159.1 NCKU01001272 RWS12576.1 NCKV01003647 RWS25498.1 GCES01004922 JAR81401.1 RQTK01001021 RUS72771.1 JH818826 EKC20634.1 HACG01005666 CEK52531.1 CP026258 AWP15880.1 AERX01026207 AERX01026208 GCES01036822 JAR49501.1 HADY01008176 HAEJ01003837 SBS44294.1 HAEB01007150 HAEC01011052 SBQ53677.1 HAED01015857 HAEE01004833 SBR02302.1 HAEH01007323 HAEI01008754 SBS05894.1 HAEF01014457 HAEG01004018 SBR70575.1 HADZ01011442 HAEA01005613 SBQ34093.1 HADW01015958 HADX01005981 SBP28213.1
JAT85721.1 NWSH01001364 PCG71539.1 KQ459591 KPI97085.1 KZ149932 PZC77310.1 AGBW02011326 OWR46822.1 JTDY01000133 KOB78685.1 KQ460473 KPJ14463.1 KK852433 KDR23960.1 GEDC01026821 GEDC01005346 JAS10477.1 JAS31952.1 GEDC01026116 JAS11182.1 NEVH01007395 PNF35918.1 ADTU01026592 KQ981522 KYN40614.1 KQ976504 KYM82868.1 GDIP01246214 JAI77187.1 GDIQ01166441 JAK85284.1 GDIQ01007635 JAN87102.1 QOIP01000007 RLU20930.1 KQ977771 KYN00032.1 LBMM01008979 KMQ88489.1 DS235750 EEB16319.1 GL888285 EGI63220.1 KQ978660 KYN29395.1 GDIQ01150795 JAL00931.1 GECL01002319 JAP03805.1 JR047032 AEY60309.1 KZ288301 PBC28845.1 AK417190 BAN20405.1 KQ414657 KOC65786.1 GECZ01016689 JAS53080.1 GECZ01028274 JAS41495.1 GEBQ01024942 JAT15035.1 GEBQ01017289 JAT22688.1 GEBQ01023820 JAT16157.1 GDKW01002508 JAI54087.1 ACPB03021239 GAHY01002192 JAA75318.1 GDIQ01134104 JAL17622.1 GDIP01209451 JAJ13951.1 GBHO01004388 GBRD01012714 JAG39216.1 JAG53110.1 NNAY01001090 OXU25174.1 KK854153 PTY12259.1 GL439158 EFN67703.1 KQ435710 KOX79700.1 KK112616 KFM58101.1 AAGJ04113373 GACK01007436 JAA57598.1 GFPF01009691 MAA20837.1 GEDV01009272 JAP79285.1 GFPF01009692 MAA20838.1 GEFH01002317 JAP66264.1 KK107390 EZA51436.1 KB202719 ESO88232.1 ABJB010182666 ABJB011105958 ABJB011127826 DS863069 EEC14182.1 GADI01002358 JAA71450.1 GFJQ02007166 JAV99803.1 KQ427052 KOF67464.1 GFWV01020320 MAA45048.1 GFAA01000276 JAU03159.1 GBBM01001845 JAC33573.1 JO844346 AEO35963.1 GBZX01001581 JAG91159.1 NCKU01001272 RWS12576.1 NCKV01003647 RWS25498.1 GCES01004922 JAR81401.1 RQTK01001021 RUS72771.1 JH818826 EKC20634.1 HACG01005666 CEK52531.1 CP026258 AWP15880.1 AERX01026207 AERX01026208 GCES01036822 JAR49501.1 HADY01008176 HAEJ01003837 SBS44294.1 HAEB01007150 HAEC01011052 SBQ53677.1 HAED01015857 HAEE01004833 SBR02302.1 HAEH01007323 HAEI01008754 SBS05894.1 HAEF01014457 HAEG01004018 SBR70575.1 HADZ01011442 HAEA01005613 SBQ34093.1 HADW01015958 HADX01005981 SBP28213.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000007151
UP000037510
+ More
UP000053240 UP000027135 UP000235965 UP000005205 UP000078541 UP000078540 UP000279307 UP000078542 UP000036403 UP000009046 UP000007755 UP000078492 UP000005203 UP000242457 UP000053825 UP000015103 UP000215335 UP000002358 UP000000311 UP000053105 UP000054359 UP000007110 UP000053097 UP000030746 UP000001555 UP000053454 UP000076420 UP000285301 UP000288716 UP000265020 UP000271974 UP000005408 UP000265000 UP000257200 UP000001038 UP000246464 UP000265160 UP000085678 UP000257160 UP000005207 UP000265100 UP000261400 UP000265080 UP000265180 UP000265200 UP000261640 UP000264800 UP000261340 UP000261600
UP000053240 UP000027135 UP000235965 UP000005205 UP000078541 UP000078540 UP000279307 UP000078542 UP000036403 UP000009046 UP000007755 UP000078492 UP000005203 UP000242457 UP000053825 UP000015103 UP000215335 UP000002358 UP000000311 UP000053105 UP000054359 UP000007110 UP000053097 UP000030746 UP000001555 UP000053454 UP000076420 UP000285301 UP000288716 UP000265020 UP000271974 UP000005408 UP000265000 UP000257200 UP000001038 UP000246464 UP000265160 UP000085678 UP000257160 UP000005207 UP000265100 UP000261400 UP000265080 UP000265180 UP000265200 UP000261640 UP000264800 UP000261340 UP000261600
PRIDE
Interpro
SUPFAM
SSF47616
SSF47616
Gene 3D
ProteinModelPortal
H9JX18
A0A3S2LUN0
A0A2H1WA37
A0A1E1WFS7
A0A2A4JIB5
A0A194Q110
+ More
A0A2W1BWX3 A0A212EZA5 A0A0L7LTA3 A0A194R9E4 A0A067RT21 A0A1B6E1Z4 A0A1B6CCM3 A0A2J7R506 A0A158NUL7 A0A195FKU3 A0A195BE71 A0A0P4X8H2 A0A0P5LYQ4 A0A0P6I9M2 A0A3L8DKH7 A0A151IFW2 A0A0J7KE41 E0VSG3 F4WRE9 A0A151JQ89 A0A088AFM0 A0A0N8BRY3 A0A0V0G761 V9IIZ0 A0A2A3EAU6 R4WIF9 A0A0L7R4T6 A0A1B6FSB1 A0A1B6EU28 A0A1B6KUB2 A0A1B6LGD4 A0A1B6KXI1 A0A0P4VJ10 R4G7S9 A0A0P5P8L6 A0A0P4ZUJ5 A0A0A9Z5R4 A0A232F340 K7J7T9 A0A2R7VY52 E2AFV4 A0A0M9A925 A0A087SZ12 W4YKT9 L7M3A2 A0A224YTA5 A0A131YL76 A0A224YT94 A0A131XFI5 A0A026W739 V3ZV28 B7Q5R0 A0A0K8RK28 A0A1Z5KY75 A0A0L8FS04 A0A293MYN1 A0A1E1XV46 A0A023GKJ7 G3MR45 A0A2C9LN35 A0A0C9S2E2 A0A443RBF0 A0A443SDA2 A0A3Q2DYX5 A0A147ASI4 A0A3S0ZDW7 K1PP76 A0A3Q2R0R9 A0A3Q1FVF1 H2MQS3 A0A0B6Y9N5 A0A2U9CIL0 A0A3P9CLF7 A0A1S3HHX9 A0A3Q1D8P2 I3J1F8 A0A146Y5N2 A0A3P8PLA0 A0A3B5ARS9 A0A1A8U7K9 A0A1A8F3H6 A0A3P8TWP4 A0A3P9KM60 A0A1A8IY73 A0A3P9JHG0 A0A1A8RKU8 A0A1A8NNE6 A0A1A8DKQ7 A0A3Q3MLB6 A0A3Q3GZT9 A0A1A7YD69 A0A3Q0S8X7 A0A3Q3K0Y7
A0A2W1BWX3 A0A212EZA5 A0A0L7LTA3 A0A194R9E4 A0A067RT21 A0A1B6E1Z4 A0A1B6CCM3 A0A2J7R506 A0A158NUL7 A0A195FKU3 A0A195BE71 A0A0P4X8H2 A0A0P5LYQ4 A0A0P6I9M2 A0A3L8DKH7 A0A151IFW2 A0A0J7KE41 E0VSG3 F4WRE9 A0A151JQ89 A0A088AFM0 A0A0N8BRY3 A0A0V0G761 V9IIZ0 A0A2A3EAU6 R4WIF9 A0A0L7R4T6 A0A1B6FSB1 A0A1B6EU28 A0A1B6KUB2 A0A1B6LGD4 A0A1B6KXI1 A0A0P4VJ10 R4G7S9 A0A0P5P8L6 A0A0P4ZUJ5 A0A0A9Z5R4 A0A232F340 K7J7T9 A0A2R7VY52 E2AFV4 A0A0M9A925 A0A087SZ12 W4YKT9 L7M3A2 A0A224YTA5 A0A131YL76 A0A224YT94 A0A131XFI5 A0A026W739 V3ZV28 B7Q5R0 A0A0K8RK28 A0A1Z5KY75 A0A0L8FS04 A0A293MYN1 A0A1E1XV46 A0A023GKJ7 G3MR45 A0A2C9LN35 A0A0C9S2E2 A0A443RBF0 A0A443SDA2 A0A3Q2DYX5 A0A147ASI4 A0A3S0ZDW7 K1PP76 A0A3Q2R0R9 A0A3Q1FVF1 H2MQS3 A0A0B6Y9N5 A0A2U9CIL0 A0A3P9CLF7 A0A1S3HHX9 A0A3Q1D8P2 I3J1F8 A0A146Y5N2 A0A3P8PLA0 A0A3B5ARS9 A0A1A8U7K9 A0A1A8F3H6 A0A3P8TWP4 A0A3P9KM60 A0A1A8IY73 A0A3P9JHG0 A0A1A8RKU8 A0A1A8NNE6 A0A1A8DKQ7 A0A3Q3MLB6 A0A3Q3GZT9 A0A1A7YD69 A0A3Q0S8X7 A0A3Q3K0Y7
Ontologies
GO
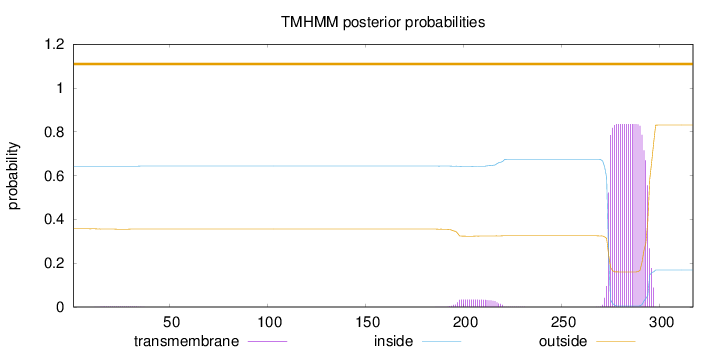
Topology
Length:
317
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
17.99665
Exp number, first 60 AAs:
0.03385
Total prob of N-in:
0.64294
outside
1 - 317
Population Genetic Test Statistics
Pi
148.897892
Theta
207.790854
Tajima's D
-0.858574
CLR
149.388894
CSRT
0.168841557922104
Interpretation
Uncertain
