Gene
KWMTBOMO12384
Pre Gene Modal
BGIBMGA014086
Annotation
endoplasmic_reticulum_protein_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.389
Sequence
CDS
ATGTCCATACAGTGGACCTTCATTGCTGGGTATCTATATTTCGAAATTGCTTTAGTGATGCTGATGATATTACCTATATTCAGTCCTAGAAGATGGAATCAATTTTTCAAATCACGTCTGTTTTCAATGTTTCGAGAAAACGTAGCTGTATATTTCTATGTGTTCATCGGTGTGTTGGCGTTGTTTTTGATTGATGCAGTGAGAGAGATACGGAAATATTCAAATGTTACGGACGTGAGCCACACTCATTTAGCTACCGAAATGAAAACTCACGTCAAGCTATTTCGAGCCCAAAGAAACTTCTATATTATAGGCTTTGCAATTTTCCTGACATTCGTAATACGTAGACTAATCACAATGTTGATTATACAAGACGAATTAAAACAGAAAGCCGAAAAAATAATAAAACAGGCAGAGGAGACAGTGAAACAGGCTAAAACAAGTATCCTAGCGAACACTTTACAATCAGAGGAATTACAACATTATGATGAAATAAATAGTCAATTAGAGGAAACTAAAATCCTATTAAAGTTAGAAAAGGATAGAGTGAAATTACTAGAGGATGATGTCAAAATGTGGCAGCAGAAATGTGAGGAAGCCATTGCCAGTAAACCCGGCAAAGGAGATGAATAA
Protein
MSIQWTFIAGYLYFEIALVMLMILPIFSPRRWNQFFKSRLFSMFRENVAVYFYVFIGVLALFLIDAVREIRKYSNVTDVSHTHLATEMKTHVKLFRAQRNFYIIGFAIFLTFVIRRLITMLIIQDELKQKAEKIIKQAEETVKQAKTSILANTLQSEELQHYDEINSQLEETKILLKLEKDRVKLLEDDVKMWQQKCEEAIASKPGKGDE
Summary
Uniprot
Q2F602
A0A1E1WAP1
A0A2A4JIK1
A0A194RAD5
A0A0L7LTZ8
A0A194PWF0
+ More
A0A2H1WBP2 S4NYA5 B4LGI7 B4L0C7 A0A0C9R1Y9 A0A212EZ42 V5FZH1 E1ZVU8 A0A1J1J0H0 A0A1W4WMV7 A0A1W4WMW9 A0A224XX42 A0A1E1WIL3 A0A023F848 A0A2A4J9Q0 S4PTH3 R4FPZ2 A0A0P4W064 A0A154PDS8 A0A2H1W8I4 A0A1B6EE10 I4DJV6 A0A232FFH5 K7J2I9 A0A0N1IGZ2 A0A1Y1MAD3 A0A194PQY2 A0A067QHE8 I4DMZ8 B9EQV3 A0A0J9RLU1 Q9W0M4 D3TRX9 B4HVK3 B4QLF9 B8A3Z7 A0A1A9ZF11 R4WPR3 A0A023FPK7 C1BQN6 A0A023FLV6 A0A023FVP8 A0A0C9QY63 U5EUL3 C1BR31 A0A151I482 A0A158NXT1 A0A146L6S9 G3MPJ8 A0A0A9WIV2 C1C264 A0A195FXC2 A0A151IT42 B7PYE7 A0A0K8RKQ7 A0A131XVN6 V5HRK4 A0A147BHU7 A0A2R7WCC4 A0A0L7QNW2
A0A2H1WBP2 S4NYA5 B4LGI7 B4L0C7 A0A0C9R1Y9 A0A212EZ42 V5FZH1 E1ZVU8 A0A1J1J0H0 A0A1W4WMV7 A0A1W4WMW9 A0A224XX42 A0A1E1WIL3 A0A023F848 A0A2A4J9Q0 S4PTH3 R4FPZ2 A0A0P4W064 A0A154PDS8 A0A2H1W8I4 A0A1B6EE10 I4DJV6 A0A232FFH5 K7J2I9 A0A0N1IGZ2 A0A1Y1MAD3 A0A194PQY2 A0A067QHE8 I4DMZ8 B9EQV3 A0A0J9RLU1 Q9W0M4 D3TRX9 B4HVK3 B4QLF9 B8A3Z7 A0A1A9ZF11 R4WPR3 A0A023FPK7 C1BQN6 A0A023FLV6 A0A023FVP8 A0A0C9QY63 U5EUL3 C1BR31 A0A151I482 A0A158NXT1 A0A146L6S9 G3MPJ8 A0A0A9WIV2 C1C264 A0A195FXC2 A0A151IT42 B7PYE7 A0A0K8RKQ7 A0A131XVN6 V5HRK4 A0A147BHU7 A0A2R7WCC4 A0A0L7QNW2
Pubmed
19121390
26354079
26227816
23622113
17994087
22118469
+ More
20798317 25474469 27129103 22651552 28648823 20075255 28004739 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 20353571 23691247 26131772 21347285 26823975 22216098 25401762 25765539 29652888
20798317 25474469 27129103 22651552 28648823 20075255 28004739 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 20353571 23691247 26131772 21347285 26823975 22216098 25401762 25765539 29652888
EMBL
BABH01042712
DQ311270
ABD36215.1
GDQN01006992
JAT84062.1
NWSH01001364
+ More
PCG71538.1 KQ460473 KPJ14464.1 JTDY01000133 KOB78686.1 KQ459591 KPI97084.1 ODYU01007280 SOQ49914.1 GAIX01013880 JAA78680.1 CH940647 EDW69425.1 CH933809 EDW18073.1 GBYB01006847 JAG76614.1 AGBW02011376 OWR46766.1 GALX01005376 JAB63090.1 GL434640 EFN74699.1 CVRI01000063 CRL04337.1 GFTR01003865 JAW12561.1 GDQN01004242 GDQN01002380 JAT86812.1 JAT88674.1 GBBI01001289 JAC17423.1 NWSH01002387 PCG68408.1 GAIX01012038 JAA80522.1 ACPB03021114 GAHY01000659 JAA76851.1 GDKW01000352 JAI56243.1 KQ434869 KZC09340.1 ODYU01007027 SOQ49409.1 GEDC01001136 JAS36162.1 AK401574 BAM18196.1 NNAY01000289 OXU29445.1 KQ459784 KPJ19953.1 GEZM01037765 JAV81948.1 KQ459595 KPI95717.1 KK853387 KDR07949.1 AK402666 BAM19288.1 BT058028 AE014296 ACM16745.1 AFH04209.1 CM002912 KMY96727.1 AY069750 AAF47419.1 AAL39895.1 AAN11453.1 EZ424181 ADD20457.1 CH480817 EDW49968.1 CM000363 EDX08732.1 KMY96726.1 BT056289 ACL68736.1 AK417656 BAN20871.1 GBBK01001884 JAC22598.1 BT076915 ACO11339.1 GBBK01001890 JAC22592.1 GBBL01001781 JAC25539.1 GBZX01003060 JAG89680.1 GANO01002270 JAB57601.1 BT077060 ACO11484.1 KQ976462 KYM84650.1 ADTU01003506 GDHC01019511 GDHC01016269 JAP99117.1 JAQ02360.1 JO843799 AEO35416.1 GBHO01035905 GBHO01035900 GBRD01011559 JAG07699.1 JAG07704.1 JAG54265.1 BT080943 ACO15367.1 KQ981204 KYN44962.1 KQ981033 KYN10174.1 ABJB010062317 ABJB010785398 ABJB010945037 DS819759 EEC11619.1 GADI01002152 JAA71656.1 GEFM01005521 JAP70275.1 GANP01007795 JAB76673.1 GEGO01005041 JAR90363.1 KK854536 PTY16660.1 KQ414843 KOC60310.1
PCG71538.1 KQ460473 KPJ14464.1 JTDY01000133 KOB78686.1 KQ459591 KPI97084.1 ODYU01007280 SOQ49914.1 GAIX01013880 JAA78680.1 CH940647 EDW69425.1 CH933809 EDW18073.1 GBYB01006847 JAG76614.1 AGBW02011376 OWR46766.1 GALX01005376 JAB63090.1 GL434640 EFN74699.1 CVRI01000063 CRL04337.1 GFTR01003865 JAW12561.1 GDQN01004242 GDQN01002380 JAT86812.1 JAT88674.1 GBBI01001289 JAC17423.1 NWSH01002387 PCG68408.1 GAIX01012038 JAA80522.1 ACPB03021114 GAHY01000659 JAA76851.1 GDKW01000352 JAI56243.1 KQ434869 KZC09340.1 ODYU01007027 SOQ49409.1 GEDC01001136 JAS36162.1 AK401574 BAM18196.1 NNAY01000289 OXU29445.1 KQ459784 KPJ19953.1 GEZM01037765 JAV81948.1 KQ459595 KPI95717.1 KK853387 KDR07949.1 AK402666 BAM19288.1 BT058028 AE014296 ACM16745.1 AFH04209.1 CM002912 KMY96727.1 AY069750 AAF47419.1 AAL39895.1 AAN11453.1 EZ424181 ADD20457.1 CH480817 EDW49968.1 CM000363 EDX08732.1 KMY96726.1 BT056289 ACL68736.1 AK417656 BAN20871.1 GBBK01001884 JAC22598.1 BT076915 ACO11339.1 GBBK01001890 JAC22592.1 GBBL01001781 JAC25539.1 GBZX01003060 JAG89680.1 GANO01002270 JAB57601.1 BT077060 ACO11484.1 KQ976462 KYM84650.1 ADTU01003506 GDHC01019511 GDHC01016269 JAP99117.1 JAQ02360.1 JO843799 AEO35416.1 GBHO01035905 GBHO01035900 GBRD01011559 JAG07699.1 JAG07704.1 JAG54265.1 BT080943 ACO15367.1 KQ981204 KYN44962.1 KQ981033 KYN10174.1 ABJB010062317 ABJB010785398 ABJB010945037 DS819759 EEC11619.1 GADI01002152 JAA71656.1 GEFM01005521 JAP70275.1 GANP01007795 JAB76673.1 GEGO01005041 JAR90363.1 KK854536 PTY16660.1 KQ414843 KOC60310.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000037510
UP000053268
UP000008792
+ More
UP000009192 UP000007151 UP000000311 UP000183832 UP000192223 UP000015103 UP000076502 UP000215335 UP000002358 UP000027135 UP000000803 UP000001292 UP000000304 UP000092445 UP000078540 UP000005205 UP000078541 UP000078492 UP000001555 UP000053825
UP000009192 UP000007151 UP000000311 UP000183832 UP000192223 UP000015103 UP000076502 UP000215335 UP000002358 UP000027135 UP000000803 UP000001292 UP000000304 UP000092445 UP000078540 UP000005205 UP000078541 UP000078492 UP000001555 UP000053825
Interpro
ProteinModelPortal
Q2F602
A0A1E1WAP1
A0A2A4JIK1
A0A194RAD5
A0A0L7LTZ8
A0A194PWF0
+ More
A0A2H1WBP2 S4NYA5 B4LGI7 B4L0C7 A0A0C9R1Y9 A0A212EZ42 V5FZH1 E1ZVU8 A0A1J1J0H0 A0A1W4WMV7 A0A1W4WMW9 A0A224XX42 A0A1E1WIL3 A0A023F848 A0A2A4J9Q0 S4PTH3 R4FPZ2 A0A0P4W064 A0A154PDS8 A0A2H1W8I4 A0A1B6EE10 I4DJV6 A0A232FFH5 K7J2I9 A0A0N1IGZ2 A0A1Y1MAD3 A0A194PQY2 A0A067QHE8 I4DMZ8 B9EQV3 A0A0J9RLU1 Q9W0M4 D3TRX9 B4HVK3 B4QLF9 B8A3Z7 A0A1A9ZF11 R4WPR3 A0A023FPK7 C1BQN6 A0A023FLV6 A0A023FVP8 A0A0C9QY63 U5EUL3 C1BR31 A0A151I482 A0A158NXT1 A0A146L6S9 G3MPJ8 A0A0A9WIV2 C1C264 A0A195FXC2 A0A151IT42 B7PYE7 A0A0K8RKQ7 A0A131XVN6 V5HRK4 A0A147BHU7 A0A2R7WCC4 A0A0L7QNW2
A0A2H1WBP2 S4NYA5 B4LGI7 B4L0C7 A0A0C9R1Y9 A0A212EZ42 V5FZH1 E1ZVU8 A0A1J1J0H0 A0A1W4WMV7 A0A1W4WMW9 A0A224XX42 A0A1E1WIL3 A0A023F848 A0A2A4J9Q0 S4PTH3 R4FPZ2 A0A0P4W064 A0A154PDS8 A0A2H1W8I4 A0A1B6EE10 I4DJV6 A0A232FFH5 K7J2I9 A0A0N1IGZ2 A0A1Y1MAD3 A0A194PQY2 A0A067QHE8 I4DMZ8 B9EQV3 A0A0J9RLU1 Q9W0M4 D3TRX9 B4HVK3 B4QLF9 B8A3Z7 A0A1A9ZF11 R4WPR3 A0A023FPK7 C1BQN6 A0A023FLV6 A0A023FVP8 A0A0C9QY63 U5EUL3 C1BR31 A0A151I482 A0A158NXT1 A0A146L6S9 G3MPJ8 A0A0A9WIV2 C1C264 A0A195FXC2 A0A151IT42 B7PYE7 A0A0K8RKQ7 A0A131XVN6 V5HRK4 A0A147BHU7 A0A2R7WCC4 A0A0L7QNW2
Ontologies
GO
PANTHER
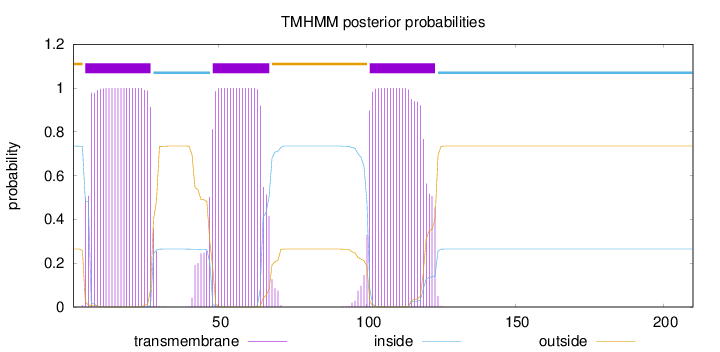
Topology
Length:
210
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
63.79162
Exp number, first 60 AAs:
36.93998
Total prob of N-in:
0.73550
POSSIBLE N-term signal
sequence
outside
1 - 4
TMhelix
5 - 27
inside
28 - 47
TMhelix
48 - 67
outside
68 - 100
TMhelix
101 - 123
inside
124 - 210
Population Genetic Test Statistics
Pi
113.730505
Theta
181.924592
Tajima's D
-1.213661
CLR
811.9491
CSRT
0.105044747762612
Interpretation
Uncertain
