Gene
KWMTBOMO12383
Annotation
PREDICTED:_ral_GTPase-activating_protein_subunit_beta-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.829 PlasmaMembrane Reliability : 1.578
Sequence
CDS
ATGAATGTGGGAATCTTCAACAGAGTTAATCATAAAGATAAAGATAGTGACGGGATGTATGCAGAATGGAGCTCAATACCACTGGAGGGTGGGGGACAAGGAGTACTGGGAGCAGTTGGAGGCCGAGATGTTGTACTGGCAGTTGTGAAACAATTGAGCTCCCCTGATCCTAGTCCTTTAACTACTGATGCTGAGGTGGAGTGGGTTCTAGAAGTGATCAGATATGGTTTGTCACTGCCACTGAGCGAGCACAGTGCTGTACGGGATTGCGTGCGTGTCGCGTGTGCTTGGTTGACCCCACTACTGGCTCCCACTCCCGCCGCGCAACCCACCCCCCCAGCTCTTCCTCCACCCGTAGTCGCCGAGCCACACAGATACGCAAGGAAGATACTGCGCCACCTACAAAACTTGTTTGTTCCAAGACCAGACGAAAGTGGCGACTTGATCAGCAAACAAGCAGTTCTGTGCCACCGCGTGCTCCGACTGGCGCGTGCTCTGGCGGACGGGGGCGCGCTGGGCGGCGCGGAGTGGAAGGCGTTGCTGTTGCTGCTGCTGGCCGCCGCTGCAGCCCTACTGGCGCCGCCGGTGCCTCCACCGCCCCCGTTCCACGCCGCAGCGATCTTTCAGCTCTGCGACCGCGTGCTCTCCGTTCTATTTGAAGTTTGGTTGGTGGCGTGTCACCGAAGTTTCCCGGGCCCCGCGCTGTGGCGTACATTGCGCGAGCAGTGCCTGCGTTGGCGGCACCGCGACCTGCTCACCAAGCATTGGACGCGCGCCTCGCTGTGCCTCACCGCGAGACTACTGAACAATCTGTATGGTCCACACTTCCCAGCCGTGCCCATCAGCGAAGAAGACGCGAACCTGGTCCCTGCGGACATGAGTCCAGAAGCGGTAATGCAAAGCTGGTATCGGATCTTGCACACGATTGGCAACCCTGTAGACTTGTGTCGACCGCGCGTCATCAGTCAAACTCCAGAATTTTTGCAGTACTCAATAACTCAAGAAGAAGGTGGTCGAGAGCCTAGCCAGCATCCATGCTTGGAAGTCCTCCCTCACATCTTTCATCAAGCAATGAAGGGTGTGGCAGCACATGTCGATGCCTTTCTAGGGCGGGGCCGCGAAGAGCGCTGGGTGTCGGGGGGCGCGGGCGGGGCGGGCGCGGGGGAGGAGCCGCCCGACGACTTCACGCCGCCCGCCCACCGCCGGCTCGCCAAGTCGTTCAGCGTCTCCGCGCCCAAACCCAAGGACAGGATACCGGAGAAAAGTACCCCTCGTACCTCCCTCATCGGCTTGAGCAGTTCGAGCAGCCGTCCACCGAGCATCGCGCCCACCACTCCGCCCAATTCTATCACTTCACAAGTGGCCGAAACGGAAAAGCCTCCCCTCACACCGTTACGTCCCAAAGTAAACTCCATCTTCCATTTATTTGGGGAGTGGCTCTTCGAGGCGGCCCTCATAGGAACCTCGGCTAACTACAACACGCAGCGTAGGCTGTGA
Protein
MNVGIFNRVNHKDKDSDGMYAEWSSIPLEGGGQGVLGAVGGRDVVLAVVKQLSSPDPSPLTTDAEVEWVLEVIRYGLSLPLSEHSAVRDCVRVACAWLTPLLAPTPAAQPTPPALPPPVVAEPHRYARKILRHLQNLFVPRPDESGDLISKQAVLCHRVLRLARALADGGALGGAEWKALLLLLLAAAAALLAPPVPPPPPFHAAAIFQLCDRVLSVLFEVWLVACHRSFPGPALWRTLREQCLRWRHRDLLTKHWTRASLCLTARLLNNLYGPHFPAVPISEEDANLVPADMSPEAVMQSWYRILHTIGNPVDLCRPRVISQTPEFLQYSITQEEGGREPSQHPCLEVLPHIFHQAMKGVAAHVDAFLGRGREERWVSGGAGGAGAGEEPPDDFTPPAHRRLAKSFSVSAPKPKDRIPEKSTPRTSLIGLSSSSSRPPSIAPTTPPNSITSQVAETEKPPLTPLRPKVNSIFHLFGEWLFEAALIGTSANYNTQRRL
Summary
Uniprot
A0A2A4JHL1
A0A2H1WA00
A0A194RB09
A0A2W1BQM4
A0A3S2L1M2
A0A194PVR4
+ More
A0A2J7PS83 A0A2J7PS81 A0A088ANE6 E2AUV9 A0A2A3ED95 A0A1B6LSA7 A0A1B6FYJ1 A0A067RK62 A0A0N1IT18 A0A0C9RA13 A0A0L7RGR3 A0A151JC06 A0A154PB09 A0A1B6MEL0 A0A1B6L443 A0A1B6D853 A0A1B6CX62 A0A158NRE5 A0A0T6AVE8 A0A212EZF5 A0A1Y1MDW0 A0A0K8S4E1 A0A0K8S613 A0A1B6MVD2 A0A0A9X2K6 D2A265 A0A1Y1MFT6 A0A1Y1MBJ0 A0A2H8TMA6 A0A232EVG5 A0A0A9ZCW2 A0A2S2QIR1 E2BVH8 A0A195FKP4 A0A2S2R3X2 A0A1B6K1C8 A0A2J7PS97 J9JWG7 N6TRH2 A0A1W4X266 A0A1W4XCZ0 A0A1W4X2A4 A0A146LGW4 A0A2H8TXZ1 A0A2H8TQC6 A0A151XI57 A0A1W4XBM7 A0A2P6L2V5 A0A151IFS4 A0A2S2NBA8 A0A2S2NQ73 T1JGQ1 U4UUE1 A0A0J7NPF2 E0VQJ7 A0A087UJA6 A0A1B6HFA9 B0W5Q0 A0A0M4DZC9 A0A0M4EDQ9 A0A1Q3FD58 A0A2M4A9E8 A0A453YTJ1 A0A195BFG5 A0A2M3Z0Z3 W5J6S7 A0A1D2NBX9 A0A2T7PG13 Q7PN19 A0A1B0D4J4 A0A0K2UWE0 A0A2M4B944 A0A2M4B8Z6 A0A2M4B917 W8B919 A0A1Q3FDF3 A0A1I8MMX3 A0A034VA45 A0A182HV79 A0A034V9G8 A0A034V7C9 A0A0A1XG88 A0A1Y9G8G0 A0A1J1INQ4 A0A182SUE6 W8AXX1 W8ALP7 A0A0L0C2G0 B4ND23 A0A240SWL3 C3ZKQ9 A0A1A9UMW0
A0A2J7PS83 A0A2J7PS81 A0A088ANE6 E2AUV9 A0A2A3ED95 A0A1B6LSA7 A0A1B6FYJ1 A0A067RK62 A0A0N1IT18 A0A0C9RA13 A0A0L7RGR3 A0A151JC06 A0A154PB09 A0A1B6MEL0 A0A1B6L443 A0A1B6D853 A0A1B6CX62 A0A158NRE5 A0A0T6AVE8 A0A212EZF5 A0A1Y1MDW0 A0A0K8S4E1 A0A0K8S613 A0A1B6MVD2 A0A0A9X2K6 D2A265 A0A1Y1MFT6 A0A1Y1MBJ0 A0A2H8TMA6 A0A232EVG5 A0A0A9ZCW2 A0A2S2QIR1 E2BVH8 A0A195FKP4 A0A2S2R3X2 A0A1B6K1C8 A0A2J7PS97 J9JWG7 N6TRH2 A0A1W4X266 A0A1W4XCZ0 A0A1W4X2A4 A0A146LGW4 A0A2H8TXZ1 A0A2H8TQC6 A0A151XI57 A0A1W4XBM7 A0A2P6L2V5 A0A151IFS4 A0A2S2NBA8 A0A2S2NQ73 T1JGQ1 U4UUE1 A0A0J7NPF2 E0VQJ7 A0A087UJA6 A0A1B6HFA9 B0W5Q0 A0A0M4DZC9 A0A0M4EDQ9 A0A1Q3FD58 A0A2M4A9E8 A0A453YTJ1 A0A195BFG5 A0A2M3Z0Z3 W5J6S7 A0A1D2NBX9 A0A2T7PG13 Q7PN19 A0A1B0D4J4 A0A0K2UWE0 A0A2M4B944 A0A2M4B8Z6 A0A2M4B917 W8B919 A0A1Q3FDF3 A0A1I8MMX3 A0A034VA45 A0A182HV79 A0A034V9G8 A0A034V7C9 A0A0A1XG88 A0A1Y9G8G0 A0A1J1INQ4 A0A182SUE6 W8AXX1 W8ALP7 A0A0L0C2G0 B4ND23 A0A240SWL3 C3ZKQ9 A0A1A9UMW0
Pubmed
EMBL
NWSH01001364
PCG71537.1
ODYU01007280
SOQ49915.1
KQ460473
KPJ14465.1
+ More
KZ149932 PZC77308.1 RSAL01000244 RVE43599.1 KQ459591 KPI97083.1 NEVH01021938 PNF19197.1 PNF19198.1 GL442898 EFN62767.1 KZ288292 PBC29189.1 GEBQ01013386 JAT26591.1 GECZ01014515 JAS55254.1 KK852424 KDR24182.1 KQ435944 KOX68194.1 GBYB01003656 JAG73423.1 KQ414596 KOC70045.1 KQ979122 KYN22519.1 KQ434864 KZC08997.1 GEBQ01005589 JAT34388.1 GEBQ01021509 JAT18468.1 GEDC01015468 GEDC01001645 JAS21830.1 JAS35653.1 GEDC01019271 GEDC01016848 JAS18027.1 JAS20450.1 ADTU01023841 ADTU01023842 LJIG01022743 KRT78948.1 AGBW02011326 OWR46824.1 GEZM01035527 JAV83198.1 GBRD01017661 JAG48166.1 GBRD01017662 JAG48165.1 GEBQ01000097 JAT39880.1 GBHO01029703 JAG13901.1 KQ971338 EFA02746.2 GEZM01035531 JAV83195.1 GEZM01035526 JAV83199.1 GFXV01001080 GFXV01003364 MBW12885.1 MBW15169.1 NNAY01002015 OXU22305.1 GBHO01001848 JAG41756.1 GGMS01008388 MBY77591.1 GL450851 EFN80302.1 KQ981522 KYN40569.1 GGMS01015420 MBY84623.1 GECU01002461 JAT05246.1 PNF19199.1 ABLF02033886 APGK01022612 KB740436 ENN80658.1 GDHC01011278 JAQ07351.1 GFXV01006984 MBW18789.1 GFXV01004540 MBW16345.1 KQ982117 KYQ59900.1 MWRG01002225 PRD32921.1 KQ977771 KYM99990.1 GGMR01001885 MBY14504.1 GGMR01006711 MBY19330.1 JH432211 KB632373 ERL93775.1 LBMM01002783 KMQ94370.1 DS235430 EEB15653.1 KK120066 KFM77445.1 GECU01034282 JAS73424.1 DS231844 EDS35703.1 CP012523 ALC37916.1 ALC38080.1 GFDL01009521 JAV25524.1 GGFK01004078 MBW37399.1 AAAB01008964 KQ976504 KYM82910.1 GGFM01001400 MBW22151.1 ADMH02001962 ETN60162.1 LJIJ01000105 ODN02585.1 PZQS01000004 PVD32368.1 EAA12904.5 AJVK01003262 AJVK01003263 AJVK01003264 AJVK01003265 AJVK01003266 HACA01025217 CDW42578.1 GGFJ01000405 MBW49546.1 GGFJ01000404 MBW49545.1 GGFJ01000403 MBW49544.1 GAMC01016884 JAB89671.1 GFDL01009465 JAV25580.1 GAKP01020524 JAC38428.1 APCN01002500 GAKP01020527 JAC38425.1 GAKP01020528 JAC38424.1 GBXI01004331 JAD09961.1 CVRI01000054 CRL00726.1 GAMC01016887 JAB89668.1 GAMC01016885 JAB89670.1 JRES01000986 KNC26436.1 CH964239 EDW82732.2 GG666638 EEN46908.1
KZ149932 PZC77308.1 RSAL01000244 RVE43599.1 KQ459591 KPI97083.1 NEVH01021938 PNF19197.1 PNF19198.1 GL442898 EFN62767.1 KZ288292 PBC29189.1 GEBQ01013386 JAT26591.1 GECZ01014515 JAS55254.1 KK852424 KDR24182.1 KQ435944 KOX68194.1 GBYB01003656 JAG73423.1 KQ414596 KOC70045.1 KQ979122 KYN22519.1 KQ434864 KZC08997.1 GEBQ01005589 JAT34388.1 GEBQ01021509 JAT18468.1 GEDC01015468 GEDC01001645 JAS21830.1 JAS35653.1 GEDC01019271 GEDC01016848 JAS18027.1 JAS20450.1 ADTU01023841 ADTU01023842 LJIG01022743 KRT78948.1 AGBW02011326 OWR46824.1 GEZM01035527 JAV83198.1 GBRD01017661 JAG48166.1 GBRD01017662 JAG48165.1 GEBQ01000097 JAT39880.1 GBHO01029703 JAG13901.1 KQ971338 EFA02746.2 GEZM01035531 JAV83195.1 GEZM01035526 JAV83199.1 GFXV01001080 GFXV01003364 MBW12885.1 MBW15169.1 NNAY01002015 OXU22305.1 GBHO01001848 JAG41756.1 GGMS01008388 MBY77591.1 GL450851 EFN80302.1 KQ981522 KYN40569.1 GGMS01015420 MBY84623.1 GECU01002461 JAT05246.1 PNF19199.1 ABLF02033886 APGK01022612 KB740436 ENN80658.1 GDHC01011278 JAQ07351.1 GFXV01006984 MBW18789.1 GFXV01004540 MBW16345.1 KQ982117 KYQ59900.1 MWRG01002225 PRD32921.1 KQ977771 KYM99990.1 GGMR01001885 MBY14504.1 GGMR01006711 MBY19330.1 JH432211 KB632373 ERL93775.1 LBMM01002783 KMQ94370.1 DS235430 EEB15653.1 KK120066 KFM77445.1 GECU01034282 JAS73424.1 DS231844 EDS35703.1 CP012523 ALC37916.1 ALC38080.1 GFDL01009521 JAV25524.1 GGFK01004078 MBW37399.1 AAAB01008964 KQ976504 KYM82910.1 GGFM01001400 MBW22151.1 ADMH02001962 ETN60162.1 LJIJ01000105 ODN02585.1 PZQS01000004 PVD32368.1 EAA12904.5 AJVK01003262 AJVK01003263 AJVK01003264 AJVK01003265 AJVK01003266 HACA01025217 CDW42578.1 GGFJ01000405 MBW49546.1 GGFJ01000404 MBW49545.1 GGFJ01000403 MBW49544.1 GAMC01016884 JAB89671.1 GFDL01009465 JAV25580.1 GAKP01020524 JAC38428.1 APCN01002500 GAKP01020527 JAC38425.1 GAKP01020528 JAC38424.1 GBXI01004331 JAD09961.1 CVRI01000054 CRL00726.1 GAMC01016887 JAB89668.1 GAMC01016885 JAB89670.1 JRES01000986 KNC26436.1 CH964239 EDW82732.2 GG666638 EEN46908.1
Proteomes
UP000218220
UP000053240
UP000283053
UP000053268
UP000235965
UP000005203
+ More
UP000000311 UP000242457 UP000027135 UP000053105 UP000053825 UP000078492 UP000076502 UP000005205 UP000007151 UP000007266 UP000215335 UP000008237 UP000078541 UP000007819 UP000019118 UP000192223 UP000075809 UP000078542 UP000030742 UP000036403 UP000009046 UP000054359 UP000002320 UP000092553 UP000078540 UP000000673 UP000094527 UP000245119 UP000007062 UP000092462 UP000095301 UP000075840 UP000069272 UP000183832 UP000075901 UP000037069 UP000007798 UP000092443 UP000001554 UP000078200
UP000000311 UP000242457 UP000027135 UP000053105 UP000053825 UP000078492 UP000076502 UP000005205 UP000007151 UP000007266 UP000215335 UP000008237 UP000078541 UP000007819 UP000019118 UP000192223 UP000075809 UP000078542 UP000030742 UP000036403 UP000009046 UP000054359 UP000002320 UP000092553 UP000078540 UP000000673 UP000094527 UP000245119 UP000007062 UP000092462 UP000095301 UP000075840 UP000069272 UP000183832 UP000075901 UP000037069 UP000007798 UP000092443 UP000001554 UP000078200
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JHL1
A0A2H1WA00
A0A194RB09
A0A2W1BQM4
A0A3S2L1M2
A0A194PVR4
+ More
A0A2J7PS83 A0A2J7PS81 A0A088ANE6 E2AUV9 A0A2A3ED95 A0A1B6LSA7 A0A1B6FYJ1 A0A067RK62 A0A0N1IT18 A0A0C9RA13 A0A0L7RGR3 A0A151JC06 A0A154PB09 A0A1B6MEL0 A0A1B6L443 A0A1B6D853 A0A1B6CX62 A0A158NRE5 A0A0T6AVE8 A0A212EZF5 A0A1Y1MDW0 A0A0K8S4E1 A0A0K8S613 A0A1B6MVD2 A0A0A9X2K6 D2A265 A0A1Y1MFT6 A0A1Y1MBJ0 A0A2H8TMA6 A0A232EVG5 A0A0A9ZCW2 A0A2S2QIR1 E2BVH8 A0A195FKP4 A0A2S2R3X2 A0A1B6K1C8 A0A2J7PS97 J9JWG7 N6TRH2 A0A1W4X266 A0A1W4XCZ0 A0A1W4X2A4 A0A146LGW4 A0A2H8TXZ1 A0A2H8TQC6 A0A151XI57 A0A1W4XBM7 A0A2P6L2V5 A0A151IFS4 A0A2S2NBA8 A0A2S2NQ73 T1JGQ1 U4UUE1 A0A0J7NPF2 E0VQJ7 A0A087UJA6 A0A1B6HFA9 B0W5Q0 A0A0M4DZC9 A0A0M4EDQ9 A0A1Q3FD58 A0A2M4A9E8 A0A453YTJ1 A0A195BFG5 A0A2M3Z0Z3 W5J6S7 A0A1D2NBX9 A0A2T7PG13 Q7PN19 A0A1B0D4J4 A0A0K2UWE0 A0A2M4B944 A0A2M4B8Z6 A0A2M4B917 W8B919 A0A1Q3FDF3 A0A1I8MMX3 A0A034VA45 A0A182HV79 A0A034V9G8 A0A034V7C9 A0A0A1XG88 A0A1Y9G8G0 A0A1J1INQ4 A0A182SUE6 W8AXX1 W8ALP7 A0A0L0C2G0 B4ND23 A0A240SWL3 C3ZKQ9 A0A1A9UMW0
A0A2J7PS83 A0A2J7PS81 A0A088ANE6 E2AUV9 A0A2A3ED95 A0A1B6LSA7 A0A1B6FYJ1 A0A067RK62 A0A0N1IT18 A0A0C9RA13 A0A0L7RGR3 A0A151JC06 A0A154PB09 A0A1B6MEL0 A0A1B6L443 A0A1B6D853 A0A1B6CX62 A0A158NRE5 A0A0T6AVE8 A0A212EZF5 A0A1Y1MDW0 A0A0K8S4E1 A0A0K8S613 A0A1B6MVD2 A0A0A9X2K6 D2A265 A0A1Y1MFT6 A0A1Y1MBJ0 A0A2H8TMA6 A0A232EVG5 A0A0A9ZCW2 A0A2S2QIR1 E2BVH8 A0A195FKP4 A0A2S2R3X2 A0A1B6K1C8 A0A2J7PS97 J9JWG7 N6TRH2 A0A1W4X266 A0A1W4XCZ0 A0A1W4X2A4 A0A146LGW4 A0A2H8TXZ1 A0A2H8TQC6 A0A151XI57 A0A1W4XBM7 A0A2P6L2V5 A0A151IFS4 A0A2S2NBA8 A0A2S2NQ73 T1JGQ1 U4UUE1 A0A0J7NPF2 E0VQJ7 A0A087UJA6 A0A1B6HFA9 B0W5Q0 A0A0M4DZC9 A0A0M4EDQ9 A0A1Q3FD58 A0A2M4A9E8 A0A453YTJ1 A0A195BFG5 A0A2M3Z0Z3 W5J6S7 A0A1D2NBX9 A0A2T7PG13 Q7PN19 A0A1B0D4J4 A0A0K2UWE0 A0A2M4B944 A0A2M4B8Z6 A0A2M4B917 W8B919 A0A1Q3FDF3 A0A1I8MMX3 A0A034VA45 A0A182HV79 A0A034V9G8 A0A034V7C9 A0A0A1XG88 A0A1Y9G8G0 A0A1J1INQ4 A0A182SUE6 W8AXX1 W8ALP7 A0A0L0C2G0 B4ND23 A0A240SWL3 C3ZKQ9 A0A1A9UMW0
Ontologies
PANTHER
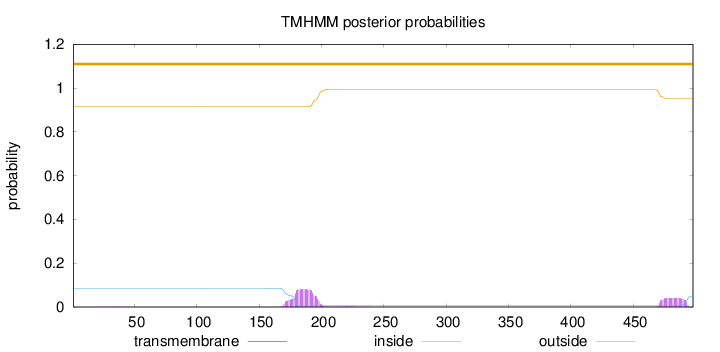
Topology
Length:
498
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.61918
Exp number, first 60 AAs:
0.00722
Total prob of N-in:
0.08554
outside
1 - 498
Population Genetic Test Statistics
Pi
183.851244
Theta
150.185048
Tajima's D
0.374493
CLR
0.06756
CSRT
0.472726363681816
Interpretation
Uncertain
