Gene
KWMTBOMO12381
Pre Gene Modal
BGIBMGA012402
Annotation
PREDICTED:_ral_GTPase-activating_protein_subunit_beta-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.65
Sequence
CDS
ATGAGTGACGCAGCGTTCCCCACGCTGGACGCCGTGCTGCGCCAGCTCGGGGACCCGGAGGCGCCGACCCTGTTCAAGCTGTTAGAGCAGCAGGCCGCCATCGAGAAACGCGTTAAAGAGAGCGAAGATAACGTGGTACCGGAAGAATATGTTCCTCCGGAGCCGGTCACAGAGTTCCAGACGGCGAGACTGTTCCTCTCGCATTTCGGCTACCTCAACATCGGAGGTGATAAAAAGGGCGGGGCGGCGCGGCTGTGGGCGCTGCGCGCGGGCGGCGCGGGCTGGGCGGCGGCGGTGCGGCGCGTGGACGCGTGCGGCGCGCGGGCGGCGCTCACCGTGCACGTGTTCTGTGCGCGCGCCGGACACACGCGCGCCGCCGACATCCTGCGCAACGCCTCCTGCACCTCCGGCCTGCCCGCCGGCTTCCTCGCCATGCTCAGAGGCCTGGGCAAACCAGTAAGGGTCCGCGGCCACGGCGGCTGGACTGGCCACGTGGACACCAGCTACGACACTCCGCATCTGCAGGATCTGCCGCCAGCCAACAACGTCGGGGAGAAAGCGCCCGCTATATACGACGGGAGGGAGCAGGTGCTGTACTGGTCGGACTCCACGAGCGAGATAGCGTTCGTGGTCCCGTCGGGGCTCGGGCCGAGCGACGCGCCGCCCGACCGCCGCAGCGACAGCATCAAGTCCGACACCGACGGGTCCTGTACATCCTCGAGGAGCTCAGAGAAGGGAGCGGGCTCGTGCTGGGACGTGTCGAGCGAGACGCACGACAAGCTGCGCGCTCTCTCGCTCGACCTGGACAGGCAGCCCGCCGCCGCGCCCGGTAGTGGACGACGTAACAGAGATTTTACTACCAAAATACTGATTATATGGCTTGAAGACTTCGAGGATCAATTGAATTTACCGATCGAGGAGCTGCTGGGCTACTGCGAGACGGGCGTGTGGTGGCGGCGCGAGGAGGTGTGCGTGGTGTGCGCGTGCGAGGCGCGCAGCGGGCTGGTGCGCGTGTCCGCCGCCGCCGCGCTGCTGGCGCCGCGCCTGCTGCCCCACGTGCTGCGCCAGCAGGCCCTCGCGCACGCGCGTCTGCACACCGACCTGTACCAGCCGCCTCACGTGAGACGGCGCCACCTCATACAGGAGCTGGCGCAGCAGTTCCGCCAGGACCTCACCGAGCCCGAGCTGCTGGAGTCGCTGTTCAAGAGTCCGTAG
Protein
MSDAAFPTLDAVLRQLGDPEAPTLFKLLEQQAAIEKRVKESEDNVVPEEYVPPEPVTEFQTARLFLSHFGYLNIGGDKKGGAARLWALRAGGAGWAAAVRRVDACGARAALTVHVFCARAGHTRAADILRNASCTSGLPAGFLAMLRGLGKPVRVRGHGGWTGHVDTSYDTPHLQDLPPANNVGEKAPAIYDGREQVLYWSDSTSEIAFVVPSGLGPSDAPPDRRSDSIKSDTDGSCTSSRSSEKGAGSCWDVSSETHDKLRALSLDLDRQPAAAPGSGRRNRDFTTKILIIWLEDFEDQLNLPIEELLGYCETGVWWRREEVCVVCACEARSGLVRVSAAAALLAPRLLPHVLRQQALAHARLHTDLYQPPHVRRRHLIQELAQQFRQDLTEPELLESLFKSP
Summary
Uniprot
H9JS92
A0A3S2L1M2
A0A2A4JHL1
A0A194PUT5
A0A212EZF5
A0A1E1WD73
+ More
A0A2W1BQM4 D2A265 A0A1W4XBM7 A0A1W4X2A4 A0A1W4XCZ0 A0A1W4X266 F4WB01 A0A0C9RA13 A0A195BFG5 A0A158NRE5 A0A1B0CD12 A0A195FKP4 A0A067RK62 A0A151IFS4 A0A026W5S6 A0A154PB09 E9IS00 A0A151JC06 E2B8D2 U4UUE1 A0A2A3ED95 A0A1B6GN48 A0A1B6M9B6 A0A0M4EDQ9 A0A0M4DZC9 A0A182JDQ4 A0A182L237 A0A182NE38 A0A182HV79 A0A182W7V6 A0A088ANE6 A0A182Q2H7 A0A0K8VSU6 A0A0K8U258 A0A0K8W8N9 A0A182PJ71 A0A182WT86 A0A1S4GXG4 A0A453YTJ1 B4JD40 A0A182RBM0 Q7PN19 A0A0L7RGR3 V5I2Y1 A0A182UUU6 A0A1B0D4J4 E2AUV9 A0A232EVG5 A0A084VP33 A0A0J7KAL8 A0A151XI57 A0A182LVU3 A0A1B6D853 A0A1B6CX62 A0A0C9R6R3 A0A0L0C2G0 W8ATY3 A0A0N1IT18 A0A3Q0S5S7 T1PDB7 W8BJ97 A0A2C9KLT9 W8ALP7 W8AXX1 A0A2C9KLT1 A0A2C9KLZ0 A0A2C9KLT3 K1QFQ5 A0A1E1XM82 V5GIH4 A0A0B7AW38 A0A0B7AVF7 A0A0B7AUY0 A0A0B7AY70 A0A210QXC5 A0A3B3Z8C3 A0A2H8TCQ9 A0A2S2NQ73 A0A2H8TXZ1 A0A2H8TQC6 A0A0A1X7V4 A0A1I8M712 A0A2S2QBN4 A0A0A1XG88 A0A2S2QLZ1 V4BMK8 A0A336MB89 A0A336MJE4 A0A336MCQ7 R7TM01 A0A293M445
A0A2W1BQM4 D2A265 A0A1W4XBM7 A0A1W4X2A4 A0A1W4XCZ0 A0A1W4X266 F4WB01 A0A0C9RA13 A0A195BFG5 A0A158NRE5 A0A1B0CD12 A0A195FKP4 A0A067RK62 A0A151IFS4 A0A026W5S6 A0A154PB09 E9IS00 A0A151JC06 E2B8D2 U4UUE1 A0A2A3ED95 A0A1B6GN48 A0A1B6M9B6 A0A0M4EDQ9 A0A0M4DZC9 A0A182JDQ4 A0A182L237 A0A182NE38 A0A182HV79 A0A182W7V6 A0A088ANE6 A0A182Q2H7 A0A0K8VSU6 A0A0K8U258 A0A0K8W8N9 A0A182PJ71 A0A182WT86 A0A1S4GXG4 A0A453YTJ1 B4JD40 A0A182RBM0 Q7PN19 A0A0L7RGR3 V5I2Y1 A0A182UUU6 A0A1B0D4J4 E2AUV9 A0A232EVG5 A0A084VP33 A0A0J7KAL8 A0A151XI57 A0A182LVU3 A0A1B6D853 A0A1B6CX62 A0A0C9R6R3 A0A0L0C2G0 W8ATY3 A0A0N1IT18 A0A3Q0S5S7 T1PDB7 W8BJ97 A0A2C9KLT9 W8ALP7 W8AXX1 A0A2C9KLT1 A0A2C9KLZ0 A0A2C9KLT3 K1QFQ5 A0A1E1XM82 V5GIH4 A0A0B7AW38 A0A0B7AVF7 A0A0B7AUY0 A0A0B7AY70 A0A210QXC5 A0A3B3Z8C3 A0A2H8TCQ9 A0A2S2NQ73 A0A2H8TXZ1 A0A2H8TQC6 A0A0A1X7V4 A0A1I8M712 A0A2S2QBN4 A0A0A1XG88 A0A2S2QLZ1 V4BMK8 A0A336MB89 A0A336MJE4 A0A336MCQ7 R7TM01 A0A293M445
Pubmed
EMBL
BABH01041314
BABH01041315
BABH01041316
BABH01041317
RSAL01000244
RVE43599.1
+ More
NWSH01001364 PCG71537.1 KQ459591 KPI97082.1 AGBW02011326 OWR46824.1 GDQN01006124 JAT84930.1 KZ149932 PZC77308.1 KQ971338 EFA02746.2 GL888056 EGI68621.1 GBYB01003656 JAG73423.1 KQ976504 KYM82910.1 ADTU01023841 ADTU01023842 AJWK01007295 KQ981522 KYN40569.1 KK852424 KDR24182.1 KQ977771 KYM99990.1 KK107390 EZA51427.1 KQ434864 KZC08997.1 GL765283 EFZ16701.1 KQ979122 KYN22519.1 GL446310 EFN88052.1 KB632373 ERL93775.1 KZ288292 PBC29189.1 GECZ01005910 JAS63859.1 GEBQ01007460 JAT32517.1 CP012523 ALC38080.1 ALC37916.1 APCN01002500 AXCN02000872 GDHF01010370 JAI41944.1 GDHF01033271 GDHF01031713 JAI19043.1 JAI20601.1 GDHF01018599 GDHF01004803 JAI33715.1 JAI47511.1 AAAB01008964 CH916368 EDW04284.1 EAA12904.5 KQ414596 KOC70045.1 GANP01002365 JAB82103.1 AJVK01003262 AJVK01003263 AJVK01003264 AJVK01003265 AJVK01003266 GL442898 EFN62767.1 NNAY01002015 OXU22305.1 ATLV01014972 KE524999 KFB39727.1 LBMM01010650 KMQ87337.1 KQ982117 KYQ59900.1 AXCM01007613 GEDC01015468 GEDC01001645 JAS21830.1 JAS35653.1 GEDC01019271 GEDC01016848 JAS18027.1 JAS20450.1 GBYB01003655 JAG73422.1 JRES01000986 KNC26436.1 GAMC01016888 JAB89667.1 KQ435944 KOX68194.1 KA646782 AFP61411.1 GAMC01016886 JAB89669.1 GAMC01016885 JAB89670.1 GAMC01016887 JAB89668.1 JH816923 EKC29924.1 GFAA01003079 JAU00356.1 GANP01014443 JAB70025.1 HACG01037982 CEK84847.1 HACG01037983 CEK84848.1 HACG01037984 CEK84849.1 HACG01037980 CEK84845.1 NEDP02001369 OWF53364.1 GFXV01000089 MBW11894.1 GGMR01006711 MBY19330.1 GFXV01006984 MBW18789.1 GFXV01004540 MBW16345.1 GBXI01006923 JAD07369.1 GGMS01005951 MBY75154.1 GBXI01004331 JAD09961.1 GGMS01009490 MBY78693.1 KB202481 ESO90184.1 UFQT01000655 SSX26189.1 SSX26188.1 SSX26187.1 AMQN01012096 KB309293 ELT94828.1 GFWV01010142 MAA34871.1
NWSH01001364 PCG71537.1 KQ459591 KPI97082.1 AGBW02011326 OWR46824.1 GDQN01006124 JAT84930.1 KZ149932 PZC77308.1 KQ971338 EFA02746.2 GL888056 EGI68621.1 GBYB01003656 JAG73423.1 KQ976504 KYM82910.1 ADTU01023841 ADTU01023842 AJWK01007295 KQ981522 KYN40569.1 KK852424 KDR24182.1 KQ977771 KYM99990.1 KK107390 EZA51427.1 KQ434864 KZC08997.1 GL765283 EFZ16701.1 KQ979122 KYN22519.1 GL446310 EFN88052.1 KB632373 ERL93775.1 KZ288292 PBC29189.1 GECZ01005910 JAS63859.1 GEBQ01007460 JAT32517.1 CP012523 ALC38080.1 ALC37916.1 APCN01002500 AXCN02000872 GDHF01010370 JAI41944.1 GDHF01033271 GDHF01031713 JAI19043.1 JAI20601.1 GDHF01018599 GDHF01004803 JAI33715.1 JAI47511.1 AAAB01008964 CH916368 EDW04284.1 EAA12904.5 KQ414596 KOC70045.1 GANP01002365 JAB82103.1 AJVK01003262 AJVK01003263 AJVK01003264 AJVK01003265 AJVK01003266 GL442898 EFN62767.1 NNAY01002015 OXU22305.1 ATLV01014972 KE524999 KFB39727.1 LBMM01010650 KMQ87337.1 KQ982117 KYQ59900.1 AXCM01007613 GEDC01015468 GEDC01001645 JAS21830.1 JAS35653.1 GEDC01019271 GEDC01016848 JAS18027.1 JAS20450.1 GBYB01003655 JAG73422.1 JRES01000986 KNC26436.1 GAMC01016888 JAB89667.1 KQ435944 KOX68194.1 KA646782 AFP61411.1 GAMC01016886 JAB89669.1 GAMC01016885 JAB89670.1 GAMC01016887 JAB89668.1 JH816923 EKC29924.1 GFAA01003079 JAU00356.1 GANP01014443 JAB70025.1 HACG01037982 CEK84847.1 HACG01037983 CEK84848.1 HACG01037984 CEK84849.1 HACG01037980 CEK84845.1 NEDP02001369 OWF53364.1 GFXV01000089 MBW11894.1 GGMR01006711 MBY19330.1 GFXV01006984 MBW18789.1 GFXV01004540 MBW16345.1 GBXI01006923 JAD07369.1 GGMS01005951 MBY75154.1 GBXI01004331 JAD09961.1 GGMS01009490 MBY78693.1 KB202481 ESO90184.1 UFQT01000655 SSX26189.1 SSX26188.1 SSX26187.1 AMQN01012096 KB309293 ELT94828.1 GFWV01010142 MAA34871.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000007151
UP000007266
+ More
UP000192223 UP000007755 UP000078540 UP000005205 UP000092461 UP000078541 UP000027135 UP000078542 UP000053097 UP000076502 UP000078492 UP000008237 UP000030742 UP000242457 UP000092553 UP000075880 UP000075882 UP000075884 UP000075840 UP000075920 UP000005203 UP000075886 UP000075885 UP000076407 UP000001070 UP000075900 UP000007062 UP000053825 UP000075903 UP000092462 UP000000311 UP000215335 UP000030765 UP000036403 UP000075809 UP000075883 UP000037069 UP000053105 UP000261340 UP000076420 UP000005408 UP000242188 UP000261520 UP000095301 UP000030746 UP000014760
UP000192223 UP000007755 UP000078540 UP000005205 UP000092461 UP000078541 UP000027135 UP000078542 UP000053097 UP000076502 UP000078492 UP000008237 UP000030742 UP000242457 UP000092553 UP000075880 UP000075882 UP000075884 UP000075840 UP000075920 UP000005203 UP000075886 UP000075885 UP000076407 UP000001070 UP000075900 UP000007062 UP000053825 UP000075903 UP000092462 UP000000311 UP000215335 UP000030765 UP000036403 UP000075809 UP000075883 UP000037069 UP000053105 UP000261340 UP000076420 UP000005408 UP000242188 UP000261520 UP000095301 UP000030746 UP000014760
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JS92
A0A3S2L1M2
A0A2A4JHL1
A0A194PUT5
A0A212EZF5
A0A1E1WD73
+ More
A0A2W1BQM4 D2A265 A0A1W4XBM7 A0A1W4X2A4 A0A1W4XCZ0 A0A1W4X266 F4WB01 A0A0C9RA13 A0A195BFG5 A0A158NRE5 A0A1B0CD12 A0A195FKP4 A0A067RK62 A0A151IFS4 A0A026W5S6 A0A154PB09 E9IS00 A0A151JC06 E2B8D2 U4UUE1 A0A2A3ED95 A0A1B6GN48 A0A1B6M9B6 A0A0M4EDQ9 A0A0M4DZC9 A0A182JDQ4 A0A182L237 A0A182NE38 A0A182HV79 A0A182W7V6 A0A088ANE6 A0A182Q2H7 A0A0K8VSU6 A0A0K8U258 A0A0K8W8N9 A0A182PJ71 A0A182WT86 A0A1S4GXG4 A0A453YTJ1 B4JD40 A0A182RBM0 Q7PN19 A0A0L7RGR3 V5I2Y1 A0A182UUU6 A0A1B0D4J4 E2AUV9 A0A232EVG5 A0A084VP33 A0A0J7KAL8 A0A151XI57 A0A182LVU3 A0A1B6D853 A0A1B6CX62 A0A0C9R6R3 A0A0L0C2G0 W8ATY3 A0A0N1IT18 A0A3Q0S5S7 T1PDB7 W8BJ97 A0A2C9KLT9 W8ALP7 W8AXX1 A0A2C9KLT1 A0A2C9KLZ0 A0A2C9KLT3 K1QFQ5 A0A1E1XM82 V5GIH4 A0A0B7AW38 A0A0B7AVF7 A0A0B7AUY0 A0A0B7AY70 A0A210QXC5 A0A3B3Z8C3 A0A2H8TCQ9 A0A2S2NQ73 A0A2H8TXZ1 A0A2H8TQC6 A0A0A1X7V4 A0A1I8M712 A0A2S2QBN4 A0A0A1XG88 A0A2S2QLZ1 V4BMK8 A0A336MB89 A0A336MJE4 A0A336MCQ7 R7TM01 A0A293M445
A0A2W1BQM4 D2A265 A0A1W4XBM7 A0A1W4X2A4 A0A1W4XCZ0 A0A1W4X266 F4WB01 A0A0C9RA13 A0A195BFG5 A0A158NRE5 A0A1B0CD12 A0A195FKP4 A0A067RK62 A0A151IFS4 A0A026W5S6 A0A154PB09 E9IS00 A0A151JC06 E2B8D2 U4UUE1 A0A2A3ED95 A0A1B6GN48 A0A1B6M9B6 A0A0M4EDQ9 A0A0M4DZC9 A0A182JDQ4 A0A182L237 A0A182NE38 A0A182HV79 A0A182W7V6 A0A088ANE6 A0A182Q2H7 A0A0K8VSU6 A0A0K8U258 A0A0K8W8N9 A0A182PJ71 A0A182WT86 A0A1S4GXG4 A0A453YTJ1 B4JD40 A0A182RBM0 Q7PN19 A0A0L7RGR3 V5I2Y1 A0A182UUU6 A0A1B0D4J4 E2AUV9 A0A232EVG5 A0A084VP33 A0A0J7KAL8 A0A151XI57 A0A182LVU3 A0A1B6D853 A0A1B6CX62 A0A0C9R6R3 A0A0L0C2G0 W8ATY3 A0A0N1IT18 A0A3Q0S5S7 T1PDB7 W8BJ97 A0A2C9KLT9 W8ALP7 W8AXX1 A0A2C9KLT1 A0A2C9KLZ0 A0A2C9KLT3 K1QFQ5 A0A1E1XM82 V5GIH4 A0A0B7AW38 A0A0B7AVF7 A0A0B7AUY0 A0A0B7AY70 A0A210QXC5 A0A3B3Z8C3 A0A2H8TCQ9 A0A2S2NQ73 A0A2H8TXZ1 A0A2H8TQC6 A0A0A1X7V4 A0A1I8M712 A0A2S2QBN4 A0A0A1XG88 A0A2S2QLZ1 V4BMK8 A0A336MB89 A0A336MJE4 A0A336MCQ7 R7TM01 A0A293M445
Ontologies
GO
PANTHER
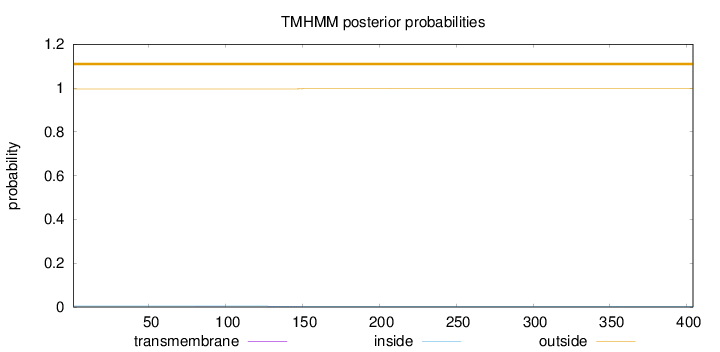
Topology
Length:
404
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0511
Exp number, first 60 AAs:
4e-05
Total prob of N-in:
0.00434
outside
1 - 404
Population Genetic Test Statistics
Pi
161.374544
Theta
135.291612
Tajima's D
-1.808136
CLR
0.356483
CSRT
0.0271986400679966
Interpretation
Uncertain
