Gene
KWMTBOMO12377
Pre Gene Modal
BGIBMGA012404
Annotation
PREDICTED:_Bloom_syndrome_protein_homolog_isoform_X1_[Bombyx_mori]
Full name
ATP-dependent DNA helicase
Location in the cell
Nuclear Reliability : 3.779
Sequence
CDS
ATGACTCCTGAGAAGATTAAATCTGACTTAAGTAATAGATTTAAGTTAAAAAACTTTATTCTGAAGAGTGACACAGGAACTCTCGATAATAAAACCAGTAAAACGAACAACAACAAAACAAAAAAGAAATTTTCTTATATAGATCCAAAAACAGCAAACAAAACATGGACACCACCATTCAAGACATTGGAGATTGTCAGTTCAGTTAAAGACGAAAACTCTCCGAAAATTCCAGAAAGATACGAAACCGGAGTCAATAGTCCTGAAAGAAGTATAGTGGTTGACATACCAACCCCACCTAAAAACGTAGACATGGACAGCTCGAGTAAAAGGAGCCGTTTGGCTATGTCTGATGACCTTAATGATGTAGATTTACGAATGAATGCCTCATTTGTACCTCTACTAGATCGAATAACAGCACATGTTGCTGTAAAGGAAACGAAACAGAAACAAAATCATCCCGAATTTCTGGATCGGCAAAGTGAAAGAATGTACGTCGAATTGTTAGAAACAATAAGTGACACGTTTGATAGAATTCCAAAGAATTTGCTAGAAATAATTCCTGGTTATGATAGTAAAATACACGACAAGATGAAACGGAGCAGAATAGAAATAAAAAAATATTGCAATAATTTTGAGAATAAATCTAGTTGCTCCATAGATAAATTACATACAACAGAGAAGGATCATAGTAATAGTTTGATACAGAATTCAACCCCGCCTTTAGAACAAAACCCTGGGACCAGTAATATGATACATTCAGATGTTAAAAAAGTCACCACTTCAAATGAATCACCGTCTGTTATGTCTGTAACCAATGATTCGACGATGGATAAAAGCTTAGATCAATCAGACACGAATGTCAAGAAGAAAGGAAAATTTGTATACAAATCCCCATTAAATCATAGCTCGCCGGATTGTATAGCTTCGTCGAGTACAGCGGAGAGACTTAAAAGCATCATGGAGACTCTGAAGCGCGTCCCACCTAAGGTTGCGTCCAATCCAACGCCGCAAGCGAATAATAGTTCAGTAGAATTCCAGCCGCCTCCGCTTGACCGAAGCGCGTTCTTGAACACGGAACGTCCGTCAACATCGGCGACACCAGCATCTGTGACTGATGTCGCCGAAAATGAATACGAACACTTGTTTGACGCCAGTGACGGCTTCGAAGAGAACATTGACCTGATTAGTGAAAATGACGTCAGCGTCGAAGAGTTTCAGAACGGTAAGAACATAAGCGTCGACGATGACGGATGGCCGGAGTACAGAATGGGGGACTTCGCGGACGACGTGGAAGTCGTTAATGCCGTCGAGATGGTGGAAATGAACTTGAACCATACGATGACGTCGAATGTCGTGAATACGAAGTACGAAGGGATCGGGCAATTCGAAGGTGATATCCAGAACGATGGAACCACAGGTGAGTTCGACGGCCTGAACTACCCTCACTCGGCTCCGATGCTGGAGATATTCCGCGAGACGTTCGGCCTGACGAAGTTCCGGCCCAACCAGCTGCAGGCCATCAACGCGGCGCTGCTCGGCCACGACTGCTTCGTCCTCATGCCGACCGGCGGCGGTAAGTCGTTGTGTTACCAGCTGCCCGCTGTACTGTCGCCCGGCGTCACCATCGTCATATCGCCGTTGAAGTCCCTTATCATGGACCAGGTCCGGAAGCTGCAGACGCTCGGCGTCCCGGCCGCCGACTTGTCCGGCAACACGTCGCTCGCAGAGACTGACGAGATCTACCACAAGCTGTCCATGAAGGAGCCGCCCATCAAGCTGCTGTACGTCACCCCCGAGAAGATCGTCGGCAGCGATCGGTTCCAGTCCGTCCTCGACTCGCTCGACGCCAGAGGGAAGTTCGCGAGGTTCGTGATAGACGAGGCGCACTGCGTGTCTCAGTGGGGGCACGAGTTCCGCCCCGACTACACGGCGCTGCGGCTGGTGCGCGCGCGCTGGCCCGCGCCCCGCGCCGTGCTGCTGACGGCCACGGCCACGCCGCGCGTGCGCCGCGACGTGCTGCTGCAGCTGCACGTGCAGCCCGCCGCCTGCCGCTGGTTCCTGTGCAGCTTCGACCGCCCCAACCTGCGCTACCGCTTCCACAAGAAACCGAAGAACATTAACCAGGAGATCGCCAAGCTCATAAAGAACGAATTTTTCGGGGAATCTGGCATAATATACTGCTTATCGTGCAAAGACTGCGAAGTGTTGGCGGGCGACCTCCACAAGACGGGGGTGCAGGCCGCCCCCTACCACGCCAGGCTCGCGGACTCCAAGCGGCAGAGCGTGCAGTCGGCCTGGATCGCCGACGAGATTAAAGTGATCTGTGCGACCATAGCGTTTGGAATGGGCATAGACAAGGCGGACGTCCGCTACGTCATGCACCACAGCATGCCCAAGTCCATAGAAGGATACTACCAAGAGTCAGGTCGCGCTGGGAGGGACGGGCAACTGGCCACCTGCATCCTGTACTACTCCTACCAGGACATGATCAGACATCGAAAATTAAACATGAGGTCGGAAATGAGGACAGAAGCGCGCAATGTCCAAGAGGAGAACCTGCGACGTATGGTGGAGCTCTGCGAGAGTGTCACTGAGTGTCGCCGCGCCTTCATCCTGGCCTACCTCGGGGAGACGCTGCCGCGCTGCAACCCTGCCCACCCTGCCGCCTGCGACAACTGTCTGCGGACACAACGCACAGAGCCGGTGGACGTGACGGAGCACTGCAAGAAGATCGTGCAGGCGGTGAGGGACGCCGGCACCCGGTCGCGGAACGGCTTCACGCTGCTGCACCTCGCGGCCGCCCTGCGCGGCTCCAAGCTGCAGCGCCTCGCCGCGCTGCACGACTCGCCCGTGCACGCACTTTGCAAGTCGTGGGACCGGAGCGACGCCCAGCGCCTGCTGCGGCTGCTGGTGGTTCGAGGGGTTCTGTACGAGAAGATCGTCGTCAACAACGACTTCGCCAGCGGATATATCGACCTGGGACCGAACGCGGACAAACTCATGAAAAATCGCATCCGCATAGTGTTCCCCATGTCGAAGACAACCGACAAGAACAGTTTAACGACAGCTCCCGCAGAGGCCCGGCCCGCGCCCCCCGCCAGCGACCTGGACTCCACGCTGGCCAAGCTCGAGCTCAGGTGCTATGCTGACCTGGTGGAAGTGTGTCGGTAA
Protein
MTPEKIKSDLSNRFKLKNFILKSDTGTLDNKTSKTNNNKTKKKFSYIDPKTANKTWTPPFKTLEIVSSVKDENSPKIPERYETGVNSPERSIVVDIPTPPKNVDMDSSSKRSRLAMSDDLNDVDLRMNASFVPLLDRITAHVAVKETKQKQNHPEFLDRQSERMYVELLETISDTFDRIPKNLLEIIPGYDSKIHDKMKRSRIEIKKYCNNFENKSSCSIDKLHTTEKDHSNSLIQNSTPPLEQNPGTSNMIHSDVKKVTTSNESPSVMSVTNDSTMDKSLDQSDTNVKKKGKFVYKSPLNHSSPDCIASSSTAERLKSIMETLKRVPPKVASNPTPQANNSSVEFQPPPLDRSAFLNTERPSTSATPASVTDVAENEYEHLFDASDGFEENIDLISENDVSVEEFQNGKNISVDDDGWPEYRMGDFADDVEVVNAVEMVEMNLNHTMTSNVVNTKYEGIGQFEGDIQNDGTTGEFDGLNYPHSAPMLEIFRETFGLTKFRPNQLQAINAALLGHDCFVLMPTGGGKSLCYQLPAVLSPGVTIVISPLKSLIMDQVRKLQTLGVPAADLSGNTSLAETDEIYHKLSMKEPPIKLLYVTPEKIVGSDRFQSVLDSLDARGKFARFVIDEAHCVSQWGHEFRPDYTALRLVRARWPAPRAVLLTATATPRVRRDVLLQLHVQPAACRWFLCSFDRPNLRYRFHKKPKNINQEIAKLIKNEFFGESGIIYCLSCKDCEVLAGDLHKTGVQAAPYHARLADSKRQSVQSAWIADEIKVICATIAFGMGIDKADVRYVMHHSMPKSIEGYYQESGRAGRDGQLATCILYYSYQDMIRHRKLNMRSEMRTEARNVQEENLRRMVELCESVTECRRAFILAYLGETLPRCNPAHPAACDNCLRTQRTEPVDVTEHCKKIVQAVRDAGTRSRNGFTLLHLAAALRGSKLQRLAALHDSPVHALCKSWDRSDAQRLLRLLVVRGVLYEKIVVNNDFASGYIDLGPNADKLMKNRIRIVFPMSKTTDKNSLTTAPAEARPAPPASDLDSTLAKLELRCYADLVEVCR
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Similarity
Belongs to the helicase family. RecQ subfamily.
Uniprot
EC Number
3.6.4.12
EMBL
Proteomes
PRIDE
Interpro
IPR032284
RecQ_Zn-bd
+ More
IPR027417 P-loop_NTPase
IPR011545 DEAD/DEAH_box_helicase_dom
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR001650 Helicase_C
IPR036390 WH_DNA-bd_sf
IPR004589 DNA_helicase_ATP-dep_RecQ
IPR014001 Helicase_ATP-bd
IPR018982 RQC_domain
IPR036388 WH-like_DNA-bd_sf
IPR010997 HRDC-like_sf
IPR002121 HRDC_dom
IPR027417 P-loop_NTPase
IPR011545 DEAD/DEAH_box_helicase_dom
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR001650 Helicase_C
IPR036390 WH_DNA-bd_sf
IPR004589 DNA_helicase_ATP-dep_RecQ
IPR014001 Helicase_ATP-bd
IPR018982 RQC_domain
IPR036388 WH-like_DNA-bd_sf
IPR010997 HRDC-like_sf
IPR002121 HRDC_dom
Gene 3D
CDD
ProteinModelPortal
PDB
4O3M
E-value=7.19036e-135,
Score=1235
Ontologies
KEGG
PATHWAY
GO
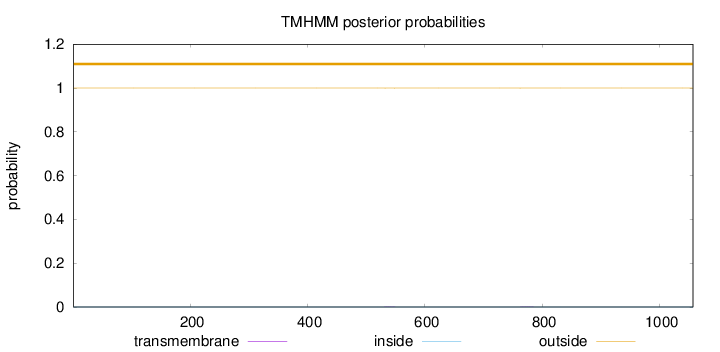
Topology
Subcellular location
Nucleus
Length:
1057
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00958
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00002
outside
1 - 1057
Population Genetic Test Statistics
Pi
181.252673
Theta
141.244067
Tajima's D
0.604071
CLR
0.668976
CSRT
0.547522623868807
Interpretation
Uncertain
