Gene
KWMTBOMO12375
Annotation
PREDICTED:_sorbitol_dehydrogenase_[Bombyx_mori]
Full name
Sorbitol dehydrogenase
Alternative Name
L-iditol 2-dehydrogenase
Polyol dehydrogenase
Xylitol dehydrogenase
Polyol dehydrogenase
Xylitol dehydrogenase
Location in the cell
Cytoplasmic Reliability : 1.719
Sequence
CDS
ATGGAGGAAGGGGCAGCGGTCCAACCGCTAGCCATCGCGATCCACGCCTGCAACCGCGCCAAGATAACTCTCGGATCTAAGATCGTTATCCTTGGAGCCGGACCTATTGGCATCTTGTGTGCTATGGCGGCCAAAGCAATGGGAGCTAGCAAAATTATTTTAACAGACGTAGTTCAGTCACGTTTAGACGCGGCACTGGAGCTGGGAGCGGATAACGTCCTCCTCGTCCGTCGGGAGTACACTGACGAAGAGGTTGTAGAAAAAATTGTGAAGTTGCTCGGTGACCGCCCGGATGTGTCAATCGATGCCTGCGGGTACGGGTCGGCGCAGAGAGTCGCTCTACTGGTGACCAAGACAGCGGGCTTGGTACTGGTGGTCGGCATAGCTGACAAAACGGTGGAGCTGCCGCTCTCACAAGCGCTGCTCAGAGAAGTTGATGTTGTAGGGTCCTTTCGTATTATGAACACGTACCAGCCCGCCCTGGCCGCGGTGTCCTCCGGGGCCATCCCCTTGGACCAGTTCATCACTCATCGCTTCCCATTGAACAAGACCAAAGAAGCACTGGATTTAGCCAAATCCGGTGCCGCCATGAAAATACTCATACACGTGCAAAATTAA
Protein
MEEGAAVQPLAIAIHACNRAKITLGSKIVILGAGPIGILCAMAAKAMGASKIILTDVVQSRLDAALELGADNVLLVRREYTDEEVVEKIVKLLGDRPDVSIDACGYGSAQRVALLVTKTAGLVLVVGIADKTVELPLSQALLREVDVVGSFRIMNTYQPALAAVSSGAIPLDQFITHRFPLNKTKEALDLAKSGAAMKILIHVQN
Summary
Description
Polyol dehydrogenase that catalyzes the reversible NAD(+)-dependent oxidation of various sugar alcohols. Is active with xylitol, L-iditol and D-sorbitol (D-glucitol) as substrates, leading to the C2-oxidized products D-xylulose, L-sorbose and D-fructose, respectively (By similarity). Is a key enzyme in the polyol pathway that interconverts glucose and fructose via sorbitol, which constitutes an important alternate route for glucose metabolism (By similarity).
Catalytic Activity
NAD(+) + xylitol = D-xylulose + H(+) + NADH
L-iditol + NAD(+) = H(+) + L-sorbose + NADH
D-fructose + H(+) + NADH = D-sorbitol + NAD(+)
L-iditol + NAD(+) = H(+) + L-sorbose + NADH
D-fructose + H(+) + NADH = D-sorbitol + NAD(+)
Cofactor
Zn(2+)
Subunit
Homotetramer.
Similarity
Belongs to the zinc-containing alcohol dehydrogenase family.
Keywords
Complete proteome
Metal-binding
NAD
Oxidoreductase
Reference proteome
Stress response
Zinc
Feature
chain Sorbitol dehydrogenase
Uniprot
Q02912
A0A2A4K6E5
A0A2W1BUJ7
A0A2H1WTR0
A0A194RA16
A0A194PUS8
+ More
A0A3S2M1Z7 A0A2A4K6T3 A0A0F6Q4H2 A0A2A4K6D8 A0A2W1BUG8 A0A3S8V4J4 A0A194R9F4 A0A1A9X1Z6 B4IVL3 A0A194PUJ4 A0A2H1WTM9 B3P2R4 B4QYY8 B4I4M2 A0A1I8P4I0 W8BR06 A0A1L8E909 A0A1A9UCP4 A0A0K8VFI9 W5JLL7 A0A0A1XLQ9 D3TNZ6 B4PSH4 A0A2M4BS11 V9IJE5 A0A1L8EFN3 A0A0K8V7I9 A0A2A3EH03 A0A1B0ACC0 O97479 Q960H1 A0A2M3Z3I5 A0A2M3ZGL7 A0A1I8NHK1 A0A1W4UU45 A0A1B0GBZ8 A0A0K8VPS5 A0A2M4BS13 H9JS89 A0A0L0BYI5 B4JV28 A0A034W7N1 B4LZX1 Q16R03 A0A2M4AK18 A0A1B6DM11 A0A0N0BII8 E9ID57 A0A2M4AJS6 A0A023ERP5 A0A1B0BKS6 A0A182H933 A0A1A9YJX5 Q1HPK9 J9WRA8 A0A0L7QMW7 Q1ET59 U5EW35 A0A2M4AIE4 A0A151XIX6 O96299 E3WI98 A0A1W4V511 B3NZK8 A0A154P6T2 A0A026VTD9 A0A1D2NEP6 B4QUM4 B4K5T2 B4PLF2 A0A3B0JSY0 A0A212EGR3 B4HIN0 F4WVG1 A0A195EP07 A0A3B0KEM2 A0A087ZU72 A0A0A1XBV0 A0A0J7L8H1 A0A195BV33 B3M0P4 A0A158NEH9 A0A3B0K615 B4NAA9 A0A1Q3FZP2 A0A0C9PQ28 A0A336K7W2 A0A1Q3FZP1 A0A437BGC5 A0A1L8E4L6 A0A1B0GIA5 A0A0P7V8S5 A0A3B0JPY2 B3LZG5
A0A3S2M1Z7 A0A2A4K6T3 A0A0F6Q4H2 A0A2A4K6D8 A0A2W1BUG8 A0A3S8V4J4 A0A194R9F4 A0A1A9X1Z6 B4IVL3 A0A194PUJ4 A0A2H1WTM9 B3P2R4 B4QYY8 B4I4M2 A0A1I8P4I0 W8BR06 A0A1L8E909 A0A1A9UCP4 A0A0K8VFI9 W5JLL7 A0A0A1XLQ9 D3TNZ6 B4PSH4 A0A2M4BS11 V9IJE5 A0A1L8EFN3 A0A0K8V7I9 A0A2A3EH03 A0A1B0ACC0 O97479 Q960H1 A0A2M3Z3I5 A0A2M3ZGL7 A0A1I8NHK1 A0A1W4UU45 A0A1B0GBZ8 A0A0K8VPS5 A0A2M4BS13 H9JS89 A0A0L0BYI5 B4JV28 A0A034W7N1 B4LZX1 Q16R03 A0A2M4AK18 A0A1B6DM11 A0A0N0BII8 E9ID57 A0A2M4AJS6 A0A023ERP5 A0A1B0BKS6 A0A182H933 A0A1A9YJX5 Q1HPK9 J9WRA8 A0A0L7QMW7 Q1ET59 U5EW35 A0A2M4AIE4 A0A151XIX6 O96299 E3WI98 A0A1W4V511 B3NZK8 A0A154P6T2 A0A026VTD9 A0A1D2NEP6 B4QUM4 B4K5T2 B4PLF2 A0A3B0JSY0 A0A212EGR3 B4HIN0 F4WVG1 A0A195EP07 A0A3B0KEM2 A0A087ZU72 A0A0A1XBV0 A0A0J7L8H1 A0A195BV33 B3M0P4 A0A158NEH9 A0A3B0K615 B4NAA9 A0A1Q3FZP2 A0A0C9PQ28 A0A336K7W2 A0A1Q3FZP1 A0A437BGC5 A0A1L8E4L6 A0A1B0GIA5 A0A0P7V8S5 A0A3B0JPY2 B3LZG5
EC Number
1.1.1.-
Pubmed
8504807
8933178
28756777
26354079
7573945
17994087
+ More
17550304 24495485 20920257 23761445 25830018 20353571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 19121390 26108605 25348373 17510324 21282665 24945155 26483478 22972362 16546206 21377527 24508170 30249741 27289101 22118469 21719571 21347285
17550304 24495485 20920257 23761445 25830018 20353571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 19121390 26108605 25348373 17510324 21282665 24945155 26483478 22972362 16546206 21377527 24508170 30249741 27289101 22118469 21719571 21347285
EMBL
D13371
D66906
NWSH01000093
PCG79639.1
KZ149932
PZC77304.1
+ More
ODYU01010939 SOQ56356.1 KQ460473 KPJ14472.1 KQ459591 KPI97077.1 RSAL01000066 RVE49388.1 PCG79628.1 KP237870 AKD01723.1 PCG79629.1 PZC77305.1 MH101955 AZL95671.1 KPJ14473.1 CH893144 EDX00549.1 KPI97076.1 SOQ56357.1 CH954181 EDV48086.1 CM000364 EDX11928.1 CH480821 EDW55165.1 GAMC01007282 GAMC01007281 JAB99273.1 GFDG01003562 JAV15237.1 GDHF01015029 JAI37285.1 ADMH02001070 ETN64198.1 GBXI01002415 JAD11877.1 EZ423148 ADD19424.1 CM000160 EDW96434.1 GGFJ01006724 MBW55865.1 JR050353 AEY61248.1 GFDG01001288 JAV17511.1 GDHF01017405 JAI34909.1 KZ288260 PBC30436.1 AF002212 AE001572 AE014297 BT044539 AAD00902.1 AAD19792.1 AAF54080.1 ACI03575.1 AHN57202.1 AY052067 AAK93491.1 GGFM01002326 MBW23077.1 GGFM01006921 MBW27672.1 CCAG010018956 GDHF01011471 JAI40843.1 GGFJ01006725 MBW55866.1 BABH01033167 JRES01001243 KNC24329.1 CH916374 EDV91348.1 GAKP01008803 GAKP01008802 JAC50150.1 CH940650 EDW67199.1 CH477727 EAT36817.1 EJY57902.1 EJY57903.1 GGFK01007800 MBW41121.1 GEDC01010583 JAS26715.1 KQ435728 KOX77804.1 GL762454 EFZ21422.1 GGFK01007547 MBW40868.1 GAPW01002237 JAC11361.1 JXJN01016005 JXUM01119833 KQ566329 KXJ70230.1 DQ443393 ABF51482.1 JX462662 AFS17318.1 KQ414868 KOC59965.1 AB164059 BAE95831.1 GANO01001600 JAB58271.1 GGFK01007230 MBW40551.1 KQ982080 KYQ60366.1 AF002213 AY058731 AAD00903.1 AAF54573.1 AAL13960.1 BABH01033168 AB604175 BAJ41475.1 EDV49581.1 KQ434814 KZC06840.1 KK107965 QOIP01000001 EZA47058.1 RLU26937.1 LJIJ01000065 ODN03731.1 EDX13431.1 CH933806 EDW15144.1 EDW96859.1 OUUW01000007 SPP83502.1 AGBW02015042 OWR40677.1 CH480815 EDW42677.1 GL888387 EGI61782.1 KQ978625 KYN29637.1 SPP83501.1 GBXI01005866 JAD08426.1 LBMM01000250 KMR02966.1 KQ976401 KYM92442.1 CH902617 EDV42059.2 ADTU01013344 ADTU01013345 ADTU01013346 SPP83500.1 CH964232 EDW81796.1 GFDL01002006 JAV33039.1 GBYB01003308 JAG73075.1 UFQS01000115 UFQS01001832 UFQT01000115 UFQT01001832 SSW99902.1 SSX12489.1 SSX20282.1 GFDL01002017 JAV33028.1 RVE49385.1 GFDF01000404 JAV13680.1 AJWK01013619 JARO02000416 KPP78637.1 OUUW01000008 SPP84185.1 EDV44144.1
ODYU01010939 SOQ56356.1 KQ460473 KPJ14472.1 KQ459591 KPI97077.1 RSAL01000066 RVE49388.1 PCG79628.1 KP237870 AKD01723.1 PCG79629.1 PZC77305.1 MH101955 AZL95671.1 KPJ14473.1 CH893144 EDX00549.1 KPI97076.1 SOQ56357.1 CH954181 EDV48086.1 CM000364 EDX11928.1 CH480821 EDW55165.1 GAMC01007282 GAMC01007281 JAB99273.1 GFDG01003562 JAV15237.1 GDHF01015029 JAI37285.1 ADMH02001070 ETN64198.1 GBXI01002415 JAD11877.1 EZ423148 ADD19424.1 CM000160 EDW96434.1 GGFJ01006724 MBW55865.1 JR050353 AEY61248.1 GFDG01001288 JAV17511.1 GDHF01017405 JAI34909.1 KZ288260 PBC30436.1 AF002212 AE001572 AE014297 BT044539 AAD00902.1 AAD19792.1 AAF54080.1 ACI03575.1 AHN57202.1 AY052067 AAK93491.1 GGFM01002326 MBW23077.1 GGFM01006921 MBW27672.1 CCAG010018956 GDHF01011471 JAI40843.1 GGFJ01006725 MBW55866.1 BABH01033167 JRES01001243 KNC24329.1 CH916374 EDV91348.1 GAKP01008803 GAKP01008802 JAC50150.1 CH940650 EDW67199.1 CH477727 EAT36817.1 EJY57902.1 EJY57903.1 GGFK01007800 MBW41121.1 GEDC01010583 JAS26715.1 KQ435728 KOX77804.1 GL762454 EFZ21422.1 GGFK01007547 MBW40868.1 GAPW01002237 JAC11361.1 JXJN01016005 JXUM01119833 KQ566329 KXJ70230.1 DQ443393 ABF51482.1 JX462662 AFS17318.1 KQ414868 KOC59965.1 AB164059 BAE95831.1 GANO01001600 JAB58271.1 GGFK01007230 MBW40551.1 KQ982080 KYQ60366.1 AF002213 AY058731 AAD00903.1 AAF54573.1 AAL13960.1 BABH01033168 AB604175 BAJ41475.1 EDV49581.1 KQ434814 KZC06840.1 KK107965 QOIP01000001 EZA47058.1 RLU26937.1 LJIJ01000065 ODN03731.1 EDX13431.1 CH933806 EDW15144.1 EDW96859.1 OUUW01000007 SPP83502.1 AGBW02015042 OWR40677.1 CH480815 EDW42677.1 GL888387 EGI61782.1 KQ978625 KYN29637.1 SPP83501.1 GBXI01005866 JAD08426.1 LBMM01000250 KMR02966.1 KQ976401 KYM92442.1 CH902617 EDV42059.2 ADTU01013344 ADTU01013345 ADTU01013346 SPP83500.1 CH964232 EDW81796.1 GFDL01002006 JAV33039.1 GBYB01003308 JAG73075.1 UFQS01000115 UFQS01001832 UFQT01000115 UFQT01001832 SSW99902.1 SSX12489.1 SSX20282.1 GFDL01002017 JAV33028.1 RVE49385.1 GFDF01000404 JAV13680.1 AJWK01013619 JARO02000416 KPP78637.1 OUUW01000008 SPP84185.1 EDV44144.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000091820
+ More
UP000002282 UP000008711 UP000000304 UP000001292 UP000095300 UP000078200 UP000000673 UP000242457 UP000092445 UP000000803 UP000095301 UP000192221 UP000092444 UP000037069 UP000001070 UP000008792 UP000008820 UP000053105 UP000092460 UP000069940 UP000249989 UP000092443 UP000053825 UP000075809 UP000076502 UP000053097 UP000279307 UP000094527 UP000009192 UP000268350 UP000007151 UP000007755 UP000078492 UP000005203 UP000036403 UP000078540 UP000007801 UP000005205 UP000007798 UP000092461 UP000034805
UP000002282 UP000008711 UP000000304 UP000001292 UP000095300 UP000078200 UP000000673 UP000242457 UP000092445 UP000000803 UP000095301 UP000192221 UP000092444 UP000037069 UP000001070 UP000008792 UP000008820 UP000053105 UP000092460 UP000069940 UP000249989 UP000092443 UP000053825 UP000075809 UP000076502 UP000053097 UP000279307 UP000094527 UP000009192 UP000268350 UP000007151 UP000007755 UP000078492 UP000005203 UP000036403 UP000078540 UP000007801 UP000005205 UP000007798 UP000092461 UP000034805
Interpro
ProteinModelPortal
Q02912
A0A2A4K6E5
A0A2W1BUJ7
A0A2H1WTR0
A0A194RA16
A0A194PUS8
+ More
A0A3S2M1Z7 A0A2A4K6T3 A0A0F6Q4H2 A0A2A4K6D8 A0A2W1BUG8 A0A3S8V4J4 A0A194R9F4 A0A1A9X1Z6 B4IVL3 A0A194PUJ4 A0A2H1WTM9 B3P2R4 B4QYY8 B4I4M2 A0A1I8P4I0 W8BR06 A0A1L8E909 A0A1A9UCP4 A0A0K8VFI9 W5JLL7 A0A0A1XLQ9 D3TNZ6 B4PSH4 A0A2M4BS11 V9IJE5 A0A1L8EFN3 A0A0K8V7I9 A0A2A3EH03 A0A1B0ACC0 O97479 Q960H1 A0A2M3Z3I5 A0A2M3ZGL7 A0A1I8NHK1 A0A1W4UU45 A0A1B0GBZ8 A0A0K8VPS5 A0A2M4BS13 H9JS89 A0A0L0BYI5 B4JV28 A0A034W7N1 B4LZX1 Q16R03 A0A2M4AK18 A0A1B6DM11 A0A0N0BII8 E9ID57 A0A2M4AJS6 A0A023ERP5 A0A1B0BKS6 A0A182H933 A0A1A9YJX5 Q1HPK9 J9WRA8 A0A0L7QMW7 Q1ET59 U5EW35 A0A2M4AIE4 A0A151XIX6 O96299 E3WI98 A0A1W4V511 B3NZK8 A0A154P6T2 A0A026VTD9 A0A1D2NEP6 B4QUM4 B4K5T2 B4PLF2 A0A3B0JSY0 A0A212EGR3 B4HIN0 F4WVG1 A0A195EP07 A0A3B0KEM2 A0A087ZU72 A0A0A1XBV0 A0A0J7L8H1 A0A195BV33 B3M0P4 A0A158NEH9 A0A3B0K615 B4NAA9 A0A1Q3FZP2 A0A0C9PQ28 A0A336K7W2 A0A1Q3FZP1 A0A437BGC5 A0A1L8E4L6 A0A1B0GIA5 A0A0P7V8S5 A0A3B0JPY2 B3LZG5
A0A3S2M1Z7 A0A2A4K6T3 A0A0F6Q4H2 A0A2A4K6D8 A0A2W1BUG8 A0A3S8V4J4 A0A194R9F4 A0A1A9X1Z6 B4IVL3 A0A194PUJ4 A0A2H1WTM9 B3P2R4 B4QYY8 B4I4M2 A0A1I8P4I0 W8BR06 A0A1L8E909 A0A1A9UCP4 A0A0K8VFI9 W5JLL7 A0A0A1XLQ9 D3TNZ6 B4PSH4 A0A2M4BS11 V9IJE5 A0A1L8EFN3 A0A0K8V7I9 A0A2A3EH03 A0A1B0ACC0 O97479 Q960H1 A0A2M3Z3I5 A0A2M3ZGL7 A0A1I8NHK1 A0A1W4UU45 A0A1B0GBZ8 A0A0K8VPS5 A0A2M4BS13 H9JS89 A0A0L0BYI5 B4JV28 A0A034W7N1 B4LZX1 Q16R03 A0A2M4AK18 A0A1B6DM11 A0A0N0BII8 E9ID57 A0A2M4AJS6 A0A023ERP5 A0A1B0BKS6 A0A182H933 A0A1A9YJX5 Q1HPK9 J9WRA8 A0A0L7QMW7 Q1ET59 U5EW35 A0A2M4AIE4 A0A151XIX6 O96299 E3WI98 A0A1W4V511 B3NZK8 A0A154P6T2 A0A026VTD9 A0A1D2NEP6 B4QUM4 B4K5T2 B4PLF2 A0A3B0JSY0 A0A212EGR3 B4HIN0 F4WVG1 A0A195EP07 A0A3B0KEM2 A0A087ZU72 A0A0A1XBV0 A0A0J7L8H1 A0A195BV33 B3M0P4 A0A158NEH9 A0A3B0K615 B4NAA9 A0A1Q3FZP2 A0A0C9PQ28 A0A336K7W2 A0A1Q3FZP1 A0A437BGC5 A0A1L8E4L6 A0A1B0GIA5 A0A0P7V8S5 A0A3B0JPY2 B3LZG5
PDB
3QE3
E-value=1.01997e-43,
Score=441
Ontologies
PATHWAY
Topology
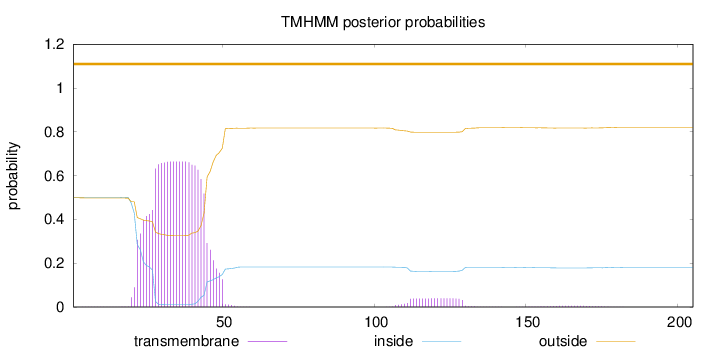
Length:
205
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
15.54771
Exp number, first 60 AAs:
14.66855
Total prob of N-in:
0.50135
POSSIBLE N-term signal
sequence
outside
1 - 205
Population Genetic Test Statistics
Pi
181.301799
Theta
135.096184
Tajima's D
0.824477
CLR
0.358717
CSRT
0.614269286535673
Interpretation
Uncertain
