Gene
KWMTBOMO12369
Pre Gene Modal
BGIBMGA012368
Annotation
PREDICTED:_bumetanide-sensitive_sodium-(potassium)-chloride_cotransporter_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.439
Sequence
CDS
ATGGGTTCCTCGCAGAAAGATATTGAATTGAGTCCGGTTCGAGGTGAAGCAAAGTCCCTGCCCGCAGATAAGCTGAACACCCAGGTACACGATGAAGAAGATGTGATAATTGGTAGATTTCGAAAGATAAGCTCAGCCAAGTCTATGGCAGCAACAAACGGGAATATTCAGTATCCCTCAGAGAGTCCTTTGCGATCTGATGATGTTGAGGCAGGACTACATTTTAATACTAAAGAGCCGTGTGAGAAGAACAAATTGCTTGATTCTGCGAACCTAGAGTTGGTGTCTCCTAAATGTTCATTAACGAAAAGTAGTTTTAGAGATGTGGAGAAACCTTCGCGGTTTAAAGATCAGCCGTCGGCTACTCGATTTCAGATGGGATACAATAAAGAAGAGGACGAGGGCACGGATGACTCCGATCCGTCGGCTATGGACAGCGACGATCCACTTACCGTATCCGATACGAAGTACGGGAAAAGTTTTAGACATTTTACACGTGAAGCTCTACCTAGATTGGACAATTACAGAAATGTTTTGTCTATACATGCCGCCCATCGACCAACTTTAGATGAGCTTCACAACGCTTCATTGTCTAGGAAGTCCGGACATACTTTAGAGAAAGACGTGGAGTCTCCGACTGTGACGTCATCGAATTTAAAGTTCGGATGGATCAAGGGCGTGCTAATGCGGTGCCTCCTGAACATATGGGGCGTGATGCTGTTCTTGAGGCTATCTTGGGTCGTGGGACAAGCTGGAGTCGCGCAGTCTGTTCTCTTGATCCTCACGACATCAGTGGTGACCACTATAACGGCCCTGTCAATGTCAGCGATCTGTACCAATGGTGTTATTAAAGGGGGTGGCACATATTACATGATATCTCGCTCATTGGGCCCAGAGTTCGGTGGTTCTATAGGTCTCATATTCTCCGTGGCTAATGCGGTGGCGTGCGCCATGTACATTGTCGGCTTCGGGGAAGCTCTGATGTCTCTCATACCAGAATCTGCTTATATGGTCAGCAAAAGTTGGGATCAAGCAATATACGGGTGCATAACGGTGGTAGTGTTGACCGGTATCGTGATAGTCGGGATGGAGTGGGAGGCCAAAGCACAGATAGTGCTGCTCATCATTCTGCTGGTGGCCATCGCGGACTTCTGCGTCGGCTCTATCATTGGCCCAAAGAGCGACGAGGAGGCCGCCCGTGGCTTCGTCGGATATAGCAGTGAGTAA
Protein
MGSSQKDIELSPVRGEAKSLPADKLNTQVHDEEDVIIGRFRKISSAKSMAATNGNIQYPSESPLRSDDVEAGLHFNTKEPCEKNKLLDSANLELVSPKCSLTKSSFRDVEKPSRFKDQPSATRFQMGYNKEEDEGTDDSDPSAMDSDDPLTVSDTKYGKSFRHFTREALPRLDNYRNVLSIHAAHRPTLDELHNASLSRKSGHTLEKDVESPTVTSSNLKFGWIKGVLMRCLLNIWGVMLFLRLSWVVGQAGVAQSVLLILTTSVVTTITALSMSAICTNGVIKGGGTYYMISRSLGPEFGGSIGLIFSVANAVACAMYIVGFGEALMSLIPESAYMVSKSWDQAIYGCITVVVLTGIVIVGMEWEAKAQIVLLIILLVAIADFCVGSIIGPKSDEEAARGFVGYSSE
Summary
Uniprot
A0A2W1BUF7
A0A3S2P0N6
A0A212F8J7
A0A194PUS3
A0A0A1WLV5
A0A0A1WPB0
+ More
A0A0P8YFC8 A0A0P8XSH9 B3M7T3 A0A0L0BWL3 A0A1I8MNN3 T1PCU7 T1PLE3 B4GRD7 A0A1I8NN16 A0A1I8NN17 A0A3B0JZA7 A0A3B0JZD4 A0A3B0K266 A0A0K8UIZ3 W8B4Z9 B4J334 W8AQQ8 A0A0R1DXB8 B4LFG0 A0A0Q9WVL2 A0A0Q9WQ97 A0A0K8WGP6 A0A034V8A2 A0A034V5K0 Q2M0N5 A0A0R3P7S7 A0A1W4UU50 A0A0R3P3H9 A0A1W4UGX5 A0A1W4UTV2 A0A0M4ELI8 A0A0R1DXN2 Q9VTW8 M9PCA7 B4PGC5 A0A0J9RUA0 B4N3I8 M9PI37 A0A0R1DXU5 A0A0J9RVB7 A0A0J9UJU1 B4HFL9 B4L068 B3NH95 A0A0Q5U446 A0A0Q5UIZ1 A0A1L8DEN0 B4QR94 A0A336LLE1 A0A182L9J3 Q8T5I9 A0A1S4GCA3 A0A182V675 A0A182WSC5 A0A182HL76 A0A182QW38 A0A1J1I274 A0A1J1HY40 A0A182N5V8 A0A182MPU2 A0A182VWK0 A0A182YAE0 A0A1Q3FV52 A0A1L8DE60 A0A182PGT7 A0A2M3YYV0 Q177B7 A0A2M3ZYG8 A0A2M4BB00 A0A182FBN2 A0A182R4U3 A0A2M4CSF9 A0A2M4CSL5 A0A2M4CSC4
A0A0P8YFC8 A0A0P8XSH9 B3M7T3 A0A0L0BWL3 A0A1I8MNN3 T1PCU7 T1PLE3 B4GRD7 A0A1I8NN16 A0A1I8NN17 A0A3B0JZA7 A0A3B0JZD4 A0A3B0K266 A0A0K8UIZ3 W8B4Z9 B4J334 W8AQQ8 A0A0R1DXB8 B4LFG0 A0A0Q9WVL2 A0A0Q9WQ97 A0A0K8WGP6 A0A034V8A2 A0A034V5K0 Q2M0N5 A0A0R3P7S7 A0A1W4UU50 A0A0R3P3H9 A0A1W4UGX5 A0A1W4UTV2 A0A0M4ELI8 A0A0R1DXN2 Q9VTW8 M9PCA7 B4PGC5 A0A0J9RUA0 B4N3I8 M9PI37 A0A0R1DXU5 A0A0J9RVB7 A0A0J9UJU1 B4HFL9 B4L068 B3NH95 A0A0Q5U446 A0A0Q5UIZ1 A0A1L8DEN0 B4QR94 A0A336LLE1 A0A182L9J3 Q8T5I9 A0A1S4GCA3 A0A182V675 A0A182WSC5 A0A182HL76 A0A182QW38 A0A1J1I274 A0A1J1HY40 A0A182N5V8 A0A182MPU2 A0A182VWK0 A0A182YAE0 A0A1Q3FV52 A0A1L8DE60 A0A182PGT7 A0A2M3YYV0 Q177B7 A0A2M3ZYG8 A0A2M4BB00 A0A182FBN2 A0A182R4U3 A0A2M4CSF9 A0A2M4CSL5 A0A2M4CSC4
Pubmed
EMBL
KZ149932
PZC77295.1
RSAL01000066
RVE49379.1
AGBW02009708
OWR50064.1
+ More
KQ459591 KPI97072.1 GBXI01014253 JAD00039.1 GBXI01013771 JAD00521.1 CH902618 KPU77647.1 KPU77645.1 EDV38806.1 KPU77646.1 JRES01001238 KNC24410.1 KA646607 AFP61236.1 KA649571 AFP64200.1 CH479188 EDW40322.1 OUUW01000012 SPP87415.1 SPP87414.1 SPP87413.1 GDHF01025642 GDHF01000020 JAI26672.1 JAI52294.1 GAMC01018179 JAB88376.1 CH916366 EDV96105.1 GAMC01018178 JAB88377.1 CM000159 KRK01744.1 CH940647 EDW69258.1 KRF84277.1 KRF84278.1 KRF84280.1 KRF84279.1 GDHF01023471 GDHF01018967 GDHF01010956 GDHF01001988 JAI28843.1 JAI33347.1 JAI41358.1 JAI50326.1 GAKP01021184 JAC37768.1 GAKP01021183 JAC37769.1 CH379069 EAL30895.2 KRT07741.1 KRT07742.1 CP012525 ALC44322.1 KRK01749.1 AE014296 AY094735 AAF49927.1 AAM11088.1 AAS65014.1 AGB94449.1 AGB94450.1 AGB94452.1 EDW94289.1 CM002912 KMY99217.1 CH964095 EDW79193.1 AGB94451.1 KRK01743.1 KRK01745.1 KRK01747.1 KRK01748.1 KMY99219.1 KMY99220.1 CH480815 EDW41250.1 CH933809 EDW18014.2 CH954178 EDV51552.1 KQS43848.1 KQS43847.1 GFDF01009165 JAV04919.1 CM000363 EDX10224.1 UFQT01000043 SSX18790.1 AJ439353 AAAB01008987 CAD27923.1 EAA00828.4 APCN01000883 AXCN02001150 CVRI01000029 CRK92473.1 CRK92474.1 AXCM01001708 GFDL01003571 JAV31474.1 GFDF01009356 JAV04728.1 GGFM01000684 MBW21435.1 CH477378 EAT42275.1 GGFK01000189 MBW33510.1 GGFJ01001059 MBW50200.1 GGFL01004079 MBW68257.1 GGFL01004077 MBW68255.1 GGFL01004078 MBW68256.1
KQ459591 KPI97072.1 GBXI01014253 JAD00039.1 GBXI01013771 JAD00521.1 CH902618 KPU77647.1 KPU77645.1 EDV38806.1 KPU77646.1 JRES01001238 KNC24410.1 KA646607 AFP61236.1 KA649571 AFP64200.1 CH479188 EDW40322.1 OUUW01000012 SPP87415.1 SPP87414.1 SPP87413.1 GDHF01025642 GDHF01000020 JAI26672.1 JAI52294.1 GAMC01018179 JAB88376.1 CH916366 EDV96105.1 GAMC01018178 JAB88377.1 CM000159 KRK01744.1 CH940647 EDW69258.1 KRF84277.1 KRF84278.1 KRF84280.1 KRF84279.1 GDHF01023471 GDHF01018967 GDHF01010956 GDHF01001988 JAI28843.1 JAI33347.1 JAI41358.1 JAI50326.1 GAKP01021184 JAC37768.1 GAKP01021183 JAC37769.1 CH379069 EAL30895.2 KRT07741.1 KRT07742.1 CP012525 ALC44322.1 KRK01749.1 AE014296 AY094735 AAF49927.1 AAM11088.1 AAS65014.1 AGB94449.1 AGB94450.1 AGB94452.1 EDW94289.1 CM002912 KMY99217.1 CH964095 EDW79193.1 AGB94451.1 KRK01743.1 KRK01745.1 KRK01747.1 KRK01748.1 KMY99219.1 KMY99220.1 CH480815 EDW41250.1 CH933809 EDW18014.2 CH954178 EDV51552.1 KQS43848.1 KQS43847.1 GFDF01009165 JAV04919.1 CM000363 EDX10224.1 UFQT01000043 SSX18790.1 AJ439353 AAAB01008987 CAD27923.1 EAA00828.4 APCN01000883 AXCN02001150 CVRI01000029 CRK92473.1 CRK92474.1 AXCM01001708 GFDL01003571 JAV31474.1 GFDF01009356 JAV04728.1 GGFM01000684 MBW21435.1 CH477378 EAT42275.1 GGFK01000189 MBW33510.1 GGFJ01001059 MBW50200.1 GGFL01004079 MBW68257.1 GGFL01004077 MBW68255.1 GGFL01004078 MBW68256.1
Proteomes
UP000283053
UP000007151
UP000053268
UP000007801
UP000037069
UP000095301
+ More
UP000008744 UP000095300 UP000268350 UP000001070 UP000002282 UP000008792 UP000001819 UP000192221 UP000092553 UP000000803 UP000007798 UP000001292 UP000009192 UP000008711 UP000000304 UP000075882 UP000007062 UP000075903 UP000076407 UP000075840 UP000075886 UP000183832 UP000075884 UP000075883 UP000075920 UP000076408 UP000075885 UP000008820 UP000069272 UP000075900
UP000008744 UP000095300 UP000268350 UP000001070 UP000002282 UP000008792 UP000001819 UP000192221 UP000092553 UP000000803 UP000007798 UP000001292 UP000009192 UP000008711 UP000000304 UP000075882 UP000007062 UP000075903 UP000076407 UP000075840 UP000075886 UP000183832 UP000075884 UP000075883 UP000075920 UP000076408 UP000075885 UP000008820 UP000069272 UP000075900
Interpro
ProteinModelPortal
A0A2W1BUF7
A0A3S2P0N6
A0A212F8J7
A0A194PUS3
A0A0A1WLV5
A0A0A1WPB0
+ More
A0A0P8YFC8 A0A0P8XSH9 B3M7T3 A0A0L0BWL3 A0A1I8MNN3 T1PCU7 T1PLE3 B4GRD7 A0A1I8NN16 A0A1I8NN17 A0A3B0JZA7 A0A3B0JZD4 A0A3B0K266 A0A0K8UIZ3 W8B4Z9 B4J334 W8AQQ8 A0A0R1DXB8 B4LFG0 A0A0Q9WVL2 A0A0Q9WQ97 A0A0K8WGP6 A0A034V8A2 A0A034V5K0 Q2M0N5 A0A0R3P7S7 A0A1W4UU50 A0A0R3P3H9 A0A1W4UGX5 A0A1W4UTV2 A0A0M4ELI8 A0A0R1DXN2 Q9VTW8 M9PCA7 B4PGC5 A0A0J9RUA0 B4N3I8 M9PI37 A0A0R1DXU5 A0A0J9RVB7 A0A0J9UJU1 B4HFL9 B4L068 B3NH95 A0A0Q5U446 A0A0Q5UIZ1 A0A1L8DEN0 B4QR94 A0A336LLE1 A0A182L9J3 Q8T5I9 A0A1S4GCA3 A0A182V675 A0A182WSC5 A0A182HL76 A0A182QW38 A0A1J1I274 A0A1J1HY40 A0A182N5V8 A0A182MPU2 A0A182VWK0 A0A182YAE0 A0A1Q3FV52 A0A1L8DE60 A0A182PGT7 A0A2M3YYV0 Q177B7 A0A2M3ZYG8 A0A2M4BB00 A0A182FBN2 A0A182R4U3 A0A2M4CSF9 A0A2M4CSL5 A0A2M4CSC4
A0A0P8YFC8 A0A0P8XSH9 B3M7T3 A0A0L0BWL3 A0A1I8MNN3 T1PCU7 T1PLE3 B4GRD7 A0A1I8NN16 A0A1I8NN17 A0A3B0JZA7 A0A3B0JZD4 A0A3B0K266 A0A0K8UIZ3 W8B4Z9 B4J334 W8AQQ8 A0A0R1DXB8 B4LFG0 A0A0Q9WVL2 A0A0Q9WQ97 A0A0K8WGP6 A0A034V8A2 A0A034V5K0 Q2M0N5 A0A0R3P7S7 A0A1W4UU50 A0A0R3P3H9 A0A1W4UGX5 A0A1W4UTV2 A0A0M4ELI8 A0A0R1DXN2 Q9VTW8 M9PCA7 B4PGC5 A0A0J9RUA0 B4N3I8 M9PI37 A0A0R1DXU5 A0A0J9RVB7 A0A0J9UJU1 B4HFL9 B4L068 B3NH95 A0A0Q5U446 A0A0Q5UIZ1 A0A1L8DEN0 B4QR94 A0A336LLE1 A0A182L9J3 Q8T5I9 A0A1S4GCA3 A0A182V675 A0A182WSC5 A0A182HL76 A0A182QW38 A0A1J1I274 A0A1J1HY40 A0A182N5V8 A0A182MPU2 A0A182VWK0 A0A182YAE0 A0A1Q3FV52 A0A1L8DE60 A0A182PGT7 A0A2M3YYV0 Q177B7 A0A2M3ZYG8 A0A2M4BB00 A0A182FBN2 A0A182R4U3 A0A2M4CSF9 A0A2M4CSL5 A0A2M4CSC4
Ontologies
GO
GO:0016021
GO:0015377
GO:0005215
GO:0006811
GO:0055085
GO:0008511
GO:0005886
GO:0015171
GO:0006813
GO:0003333
GO:0006821
GO:0005623
GO:0015379
GO:0055078
GO:0035725
GO:1902476
GO:0015081
GO:0015378
GO:0055064
GO:0055075
GO:0006884
GO:1990573
GO:0006810
GO:0016020
GO:0005524
GO:0003909
GO:0051103
GO:0000049
GO:0005975
GO:0004553
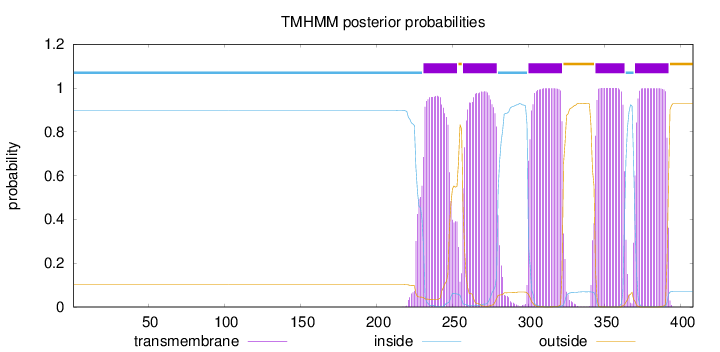
Topology
Length:
408
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
109.04117
Exp number, first 60 AAs:
0
Total prob of N-in:
0.89801
inside
1 - 230
TMhelix
231 - 253
outside
254 - 256
TMhelix
257 - 279
inside
280 - 299
TMhelix
300 - 322
outside
323 - 343
TMhelix
344 - 363
inside
364 - 369
TMhelix
370 - 392
outside
393 - 408
Population Genetic Test Statistics
Pi
212.091464
Theta
170.69299
Tajima's D
0.352264
CLR
0.19139
CSRT
0.470026498675066
Interpretation
Uncertain
