Gene
KWMTBOMO12368
Pre Gene Modal
BGIBMGA012368
Annotation
PREDICTED:_bumetanide-sensitive_sodium-(potassium)-chloride_cotransporter_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.655
Sequence
CDS
ATGTTCGTATACTTGTCTACAGTGGAGGTGCTGTGGCAGAACATGGGCCCCGACTACAGATATTACGAGGGCGTCCATCACAACTTCTTCTCGGTATTCTCCATATTCTTCCCGGCCGCTACCGGGATATTAGCCGGCGCCAACATTTCGGGAGATTTGAAGGACCCTCAGAAGTCGATTCCGAAGGGCACGCTGCTCGCCATCCTGCTGACGACGCTGTCGTACCTGCTGATAGCGGTGGTGTCGGGCTGGTCGGTGCTGCGGGACGCGTCGGGGGACGTGGCCGACCTGGCCGCCGGCTCGCTGGCCGCCTGCGTCCATAACGGTACCTGCGCGTACGGTCTGCAGAACGACTACCATGTGGTGCGGCTGGTGTCAGCCTTCGGTCCACTCATTCACGGAGGTTGCTTCGCGGCCGCGCTCTCCTCCGCGCTGGCCTCTCTGGTCTCCGCTCCGAAGGTCTTCCAGGCGCTGTGTCAGGACAAGCTGTACCCCTATATCGAGTTCTTCGCTAAGGGCTACGGACCCAACAACGAGCCCGTGCGAGGGTACGTGCTGACCTTCGTCATCGCCACCACCTTCATACTCCTGGGCGGCCTGAACCAGATAGCCCCGCTGATATCGAACTTCTTCCTGGCGGCCTACACGCTCATCAACTTCTCCACGTTCCACGCGAGCCTCGCCAGGCCCGTCGGCTGGAGACCCACCTTCAGGTTTTACAACATGTGGCTGTCGCTGCTGGGCTCGGTGCTGTGCGTGGTCATCATGTTCGTGGTGTCGTGGAGCCAGGCGCTCGGCACCTTCGCCTTCCTGCTCGCCCTCTACCTGCTCGTGTCCTACAGGAAGCCAGACGTGAACTGGGGCTCCACGACGCAGGCGCAGACGTACAAGACGGCGCTGGCCGGCGTGCAGCTGCTCAACGGGATCGACGAGCACGTCAAGAACTACAGGCCGCAAGTCCTGGTGCTGACGGGGTTCCCGGGGGAGCGGCCGCTGCTCACGGACTTCACGTACCTGCTGACCAAGGGACTGTCGCTGATGCTGTGCGGACAGATCGTTCAAGGTCCGGTGAGCCAGCCGACGCGCGAGGCCCTCAGCAAGCGAGCGTACGGCTGGTTCCAGCGCCGGAAGATCAAGGCCTTCTACACCCTGGTCGATGACGTCAGCTTCAAGGACGGCGCCAGCGCGCTGGTGCAGAGCAGCGGGCTCGGGAAGCTCACCCCCAACATCCTCATGATGGGCTTCAAGCAGGACTGGCGCACGTGCAGCAGGCAACACCTGGTGGACTACGTCGAGGTCATGCATAAAGCGTTGGACGTGCACATGGCGCTCGCGATACTGCGCATCGAGGGCGGACAGTACAGCAGCGAGGCGCTCGACGAAGACCTCCAGATCTACCTCAATGACCTCAACGCTAACGGCGTGGCGGGAATATCGCGTTCCATGGGAGATACATACAAAGCAACGAAGTCTACGGAGAGCCTCAACACTCAGTCCGAAGGCAGCACGTCTGACTTGTCGGTGGGGTCGCCACAGCTGCGACGCGCCGTCTCGCAGGACCCCGGCGCAGACCCGGACAAGGAGCAAGTCAAAATTAAAAAAAGCGGCAAAGGAAGTAAAGGTCTAAGTTACGCTGTGCGGAGGGCTCTAGGGTCTGCGGTGGCTCGCAGGGCGTCCTCGTTCACTTCAGTGGAACAATTCACGAGGAGGCAGGCCGGGACCGTCGACGTTTGGTGGCTCTATGACGACGGAGGTCTGACCCTGTTGCTGCCGTACATCCTGTCCACCAGACGCGCCTGGTCTTCCAGCCCGCTGCGGATATTCACGCTCGCCAACAAGAACACGGAGCTGGAGATCGAAGAAAGGAACATGGCGTCCCTGCTGTCCAAGTTCCGCATCGACTACTCGGCGCTGACGGTGGTGGCGGACATCACGCGGCGCCCCCGGGACTCCACGCTGGACTACTTCGACCAGCTCATCGCGCCCTTCATGGTGCCGGAAGACGCCAACCATACGGCGGGTGTGACGGCGGGAGTGACTGCGGGCGACCTGGCGGCGGCCCGCGAGCGCACGCACCGGCACCTGCGGCTGCGCGAGCTCATCGGGGAGCGGTCCCGGGGCGCGCGGCTGGTGTGCGCCACGCTGCCCATGCCGCGCCGCCGCAGCGTGGTGCCGCCGCCGCTCTACGCCGCCTGGCTGCACGCCATCGCCTCCGCCGCCGACAACACGCTGCTCGTGCGCGGCAACCACGAGCCCGTGCTCACCTTCTACTCCTGA
Protein
MFVYLSTVEVLWQNMGPDYRYYEGVHHNFFSVFSIFFPAATGILAGANISGDLKDPQKSIPKGTLLAILLTTLSYLLIAVVSGWSVLRDASGDVADLAAGSLAACVHNGTCAYGLQNDYHVVRLVSAFGPLIHGGCFAAALSSALASLVSAPKVFQALCQDKLYPYIEFFAKGYGPNNEPVRGYVLTFVIATTFILLGGLNQIAPLISNFFLAAYTLINFSTFHASLARPVGWRPTFRFYNMWLSLLGSVLCVVIMFVVSWSQALGTFAFLLALYLLVSYRKPDVNWGSTTQAQTYKTALAGVQLLNGIDEHVKNYRPQVLVLTGFPGERPLLTDFTYLLTKGLSLMLCGQIVQGPVSQPTREALSKRAYGWFQRRKIKAFYTLVDDVSFKDGASALVQSSGLGKLTPNILMMGFKQDWRTCSRQHLVDYVEVMHKALDVHMALAILRIEGGQYSSEALDEDLQIYLNDLNANGVAGISRSMGDTYKATKSTESLNTQSEGSTSDLSVGSPQLRRAVSQDPGADPDKEQVKIKKSGKGSKGLSYAVRRALGSAVARRASSFTSVEQFTRRQAGTVDVWWLYDDGGLTLLLPYILSTRRAWSSSPLRIFTLANKNTELEIEERNMASLLSKFRIDYSALTVVADITRRPRDSTLDYFDQLIAPFMVPEDANHTAGVTAGVTAGDLAAARERTHRHLRLRELIGERSRGARLVCATLPMPRRRSVVPPPLYAAWLHAIASAADNTLLVRGNHEPVLTFYS
Summary
Uniprot
H9JS58
A0A2W1BUF7
A0A2A4K7K6
A0A212F8J7
A0A2J7Q344
A0A0L0BWL3
+ More
A0A067QZ58 W8AQQ8 T1PLE3 T1PCU7 A0A1I8MNN3 W8B4Z9 A0A034V8A2 A0A034V5K0 B4J334 A0A0P8YFC8 A0A0K8WGP6 A0A0P8XSH9 B3M7T3 A0A1A9WEC2 B4L068 A0A182K428 A0A182N5V8 Q8T5I9 B4QR94 A0A0J9RVB7 A0A182TW57 A0A1I8NN16 A0A1I8NN17 A0A182HL76 A0A1W4WT68 A0A3B0JZA7 A0A1S4GCA3 Q9VTW8 A0A182WSC5 A0A3B0K266 A0A182QW38 B4PGC5 A0A1W4WH00 A0A0A9WLJ8 A0A3B0JZD4 A0A182V675 A0A182IUZ5 A0A0K8THF8 A0A0Q9WQ97 A0A0J9RUA0 B3NH95 B4LFG0 A0A182R4U3 Q2M0N5 A0A2M4CSC4 A0A2M3YYV0 A0A0R3P7S7 W5JDN1 A0A0R1DXN2 A0A2M4CSL5 A0A2M4BB00 M9PCA7 A0A182PGT7 A0A1W4UGX5 A0A2M3ZY80 A0A2M4BCB9 D2A527 A0A2M4CSF9 A0A2M3ZYG8 A0A182FBN2 A0A182MPU2 A0A0Q5U446 A0A1W4UU50 A0A182VWK0 A0A154PG76 A0A182YAE0 A0A0Q9WVL2 A0A0J9RU03 A0A1Q3FV52 A0A1Q3FYY0 A0A182GFY2 A0A0R1E2A3 A0A1B6CXS0 A0A1B6D1J5 A0A1B6D5F3 A0A0M4ELI8 B4N3I8 A0A195FZH4 A0A182L9J3 A0A1B6KU44 A0A1Y1L619 A0A195DRW0 A0A0L7R055 A0A195BTY2 A0A1B6I7U9 A0A1B6K8B8 A0A026WBV0 A0A158NVA6 A0A1B6K518 A0A0J9UJU1 M9PI37 A0A0R1DXU5 A0A0Q5UIZ1 A0A151WUM5
A0A067QZ58 W8AQQ8 T1PLE3 T1PCU7 A0A1I8MNN3 W8B4Z9 A0A034V8A2 A0A034V5K0 B4J334 A0A0P8YFC8 A0A0K8WGP6 A0A0P8XSH9 B3M7T3 A0A1A9WEC2 B4L068 A0A182K428 A0A182N5V8 Q8T5I9 B4QR94 A0A0J9RVB7 A0A182TW57 A0A1I8NN16 A0A1I8NN17 A0A182HL76 A0A1W4WT68 A0A3B0JZA7 A0A1S4GCA3 Q9VTW8 A0A182WSC5 A0A3B0K266 A0A182QW38 B4PGC5 A0A1W4WH00 A0A0A9WLJ8 A0A3B0JZD4 A0A182V675 A0A182IUZ5 A0A0K8THF8 A0A0Q9WQ97 A0A0J9RUA0 B3NH95 B4LFG0 A0A182R4U3 Q2M0N5 A0A2M4CSC4 A0A2M3YYV0 A0A0R3P7S7 W5JDN1 A0A0R1DXN2 A0A2M4CSL5 A0A2M4BB00 M9PCA7 A0A182PGT7 A0A1W4UGX5 A0A2M3ZY80 A0A2M4BCB9 D2A527 A0A2M4CSF9 A0A2M3ZYG8 A0A182FBN2 A0A182MPU2 A0A0Q5U446 A0A1W4UU50 A0A182VWK0 A0A154PG76 A0A182YAE0 A0A0Q9WVL2 A0A0J9RU03 A0A1Q3FV52 A0A1Q3FYY0 A0A182GFY2 A0A0R1E2A3 A0A1B6CXS0 A0A1B6D1J5 A0A1B6D5F3 A0A0M4ELI8 B4N3I8 A0A195FZH4 A0A182L9J3 A0A1B6KU44 A0A1Y1L619 A0A195DRW0 A0A0L7R055 A0A195BTY2 A0A1B6I7U9 A0A1B6K8B8 A0A026WBV0 A0A158NVA6 A0A1B6K518 A0A0J9UJU1 M9PI37 A0A0R1DXU5 A0A0Q5UIZ1 A0A151WUM5
Pubmed
19121390
28756777
22118469
26108605
24845553
24495485
+ More
25315136 25348373 17994087 18057021 12060762 12364791 14747013 17210077 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25401762 26823975 15632085 20920257 23761445 18362917 19820115 25244985 26483478 20966253 28004739 24508170 21347285
25315136 25348373 17994087 18057021 12060762 12364791 14747013 17210077 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25401762 26823975 15632085 20920257 23761445 18362917 19820115 25244985 26483478 20966253 28004739 24508170 21347285
EMBL
BABH01033155
BABH01033156
BABH01033157
BABH01033158
KZ149932
PZC77295.1
+ More
NWSH01000093 PCG79632.1 AGBW02009708 OWR50064.1 NEVH01019070 PNF23008.1 JRES01001238 KNC24410.1 KK853171 KDR10326.1 GAMC01018178 JAB88377.1 KA649571 AFP64200.1 KA646607 AFP61236.1 GAMC01018179 JAB88376.1 GAKP01021184 JAC37768.1 GAKP01021183 JAC37769.1 CH916366 EDV96105.1 CH902618 KPU77647.1 GDHF01023471 GDHF01018967 GDHF01010956 GDHF01001988 JAI28843.1 JAI33347.1 JAI41358.1 JAI50326.1 KPU77645.1 EDV38806.1 KPU77646.1 CH933809 EDW18014.2 AJ439353 AAAB01008987 CAD27923.1 EAA00828.4 CM000363 EDX10224.1 CM002912 KMY99219.1 APCN01000883 OUUW01000012 SPP87415.1 AE014296 AY094735 AAF49927.1 AAM11088.1 AAS65014.1 SPP87413.1 AXCN02001150 CM000159 EDW94289.1 GBHO01037874 GBHO01006440 GBHO01006439 GDHC01017468 JAG05730.1 JAG37164.1 JAG37165.1 JAQ01161.1 SPP87414.1 GBRD01000867 JAG64954.1 CH940647 KRF84279.1 KMY99217.1 CH954178 EDV51552.1 EDW69258.1 CH379069 EAL30895.2 GGFL01004078 MBW68256.1 GGFM01000684 MBW21435.1 KRT07741.1 ADMH02001683 ETN61443.1 KRK01749.1 GGFL01004077 MBW68255.1 GGFJ01001059 MBW50200.1 AGB94449.1 AGB94450.1 AGB94452.1 GGFK01000205 MBW33526.1 GGFJ01001558 MBW50699.1 KQ971361 EFA05305.2 GGFL01004079 MBW68257.1 GGFK01000189 MBW33510.1 AXCM01001708 KQS43848.1 KQ434889 KZC10308.1 KRF84277.1 KRF84278.1 KRF84280.1 KMY99218.1 GFDL01003571 JAV31474.1 GFDL01002302 JAV32743.1 JXUM01010846 JXUM01010847 JXUM01010848 JXUM01010849 KQ560318 KXJ83080.1 KRK01746.1 GEDC01019031 JAS18267.1 GEDC01017805 JAS19493.1 GEDC01016379 JAS20919.1 CP012525 ALC44322.1 CH964095 EDW79193.1 KQ981193 KYN45204.1 GEBQ01025056 JAT14921.1 GEZM01065969 JAV68258.1 KQ980530 KYN15608.1 KQ414672 KOC64235.1 KQ976417 KYM89879.1 GECU01024714 JAS82992.1 GECU01000014 JAT07693.1 KK107324 EZA52534.1 ADTU01027052 ADTU01027053 GECU01001443 JAT06264.1 KMY99220.1 AGB94451.1 KRK01743.1 KRK01745.1 KRK01747.1 KRK01748.1 KQS43847.1 KQ982745 KYQ51345.1
NWSH01000093 PCG79632.1 AGBW02009708 OWR50064.1 NEVH01019070 PNF23008.1 JRES01001238 KNC24410.1 KK853171 KDR10326.1 GAMC01018178 JAB88377.1 KA649571 AFP64200.1 KA646607 AFP61236.1 GAMC01018179 JAB88376.1 GAKP01021184 JAC37768.1 GAKP01021183 JAC37769.1 CH916366 EDV96105.1 CH902618 KPU77647.1 GDHF01023471 GDHF01018967 GDHF01010956 GDHF01001988 JAI28843.1 JAI33347.1 JAI41358.1 JAI50326.1 KPU77645.1 EDV38806.1 KPU77646.1 CH933809 EDW18014.2 AJ439353 AAAB01008987 CAD27923.1 EAA00828.4 CM000363 EDX10224.1 CM002912 KMY99219.1 APCN01000883 OUUW01000012 SPP87415.1 AE014296 AY094735 AAF49927.1 AAM11088.1 AAS65014.1 SPP87413.1 AXCN02001150 CM000159 EDW94289.1 GBHO01037874 GBHO01006440 GBHO01006439 GDHC01017468 JAG05730.1 JAG37164.1 JAG37165.1 JAQ01161.1 SPP87414.1 GBRD01000867 JAG64954.1 CH940647 KRF84279.1 KMY99217.1 CH954178 EDV51552.1 EDW69258.1 CH379069 EAL30895.2 GGFL01004078 MBW68256.1 GGFM01000684 MBW21435.1 KRT07741.1 ADMH02001683 ETN61443.1 KRK01749.1 GGFL01004077 MBW68255.1 GGFJ01001059 MBW50200.1 AGB94449.1 AGB94450.1 AGB94452.1 GGFK01000205 MBW33526.1 GGFJ01001558 MBW50699.1 KQ971361 EFA05305.2 GGFL01004079 MBW68257.1 GGFK01000189 MBW33510.1 AXCM01001708 KQS43848.1 KQ434889 KZC10308.1 KRF84277.1 KRF84278.1 KRF84280.1 KMY99218.1 GFDL01003571 JAV31474.1 GFDL01002302 JAV32743.1 JXUM01010846 JXUM01010847 JXUM01010848 JXUM01010849 KQ560318 KXJ83080.1 KRK01746.1 GEDC01019031 JAS18267.1 GEDC01017805 JAS19493.1 GEDC01016379 JAS20919.1 CP012525 ALC44322.1 CH964095 EDW79193.1 KQ981193 KYN45204.1 GEBQ01025056 JAT14921.1 GEZM01065969 JAV68258.1 KQ980530 KYN15608.1 KQ414672 KOC64235.1 KQ976417 KYM89879.1 GECU01024714 JAS82992.1 GECU01000014 JAT07693.1 KK107324 EZA52534.1 ADTU01027052 ADTU01027053 GECU01001443 JAT06264.1 KMY99220.1 AGB94451.1 KRK01743.1 KRK01745.1 KRK01747.1 KRK01748.1 KQS43847.1 KQ982745 KYQ51345.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000235965
UP000037069
UP000027135
+ More
UP000095301 UP000001070 UP000007801 UP000091820 UP000009192 UP000075881 UP000075884 UP000007062 UP000000304 UP000075902 UP000095300 UP000075840 UP000192223 UP000268350 UP000000803 UP000076407 UP000075886 UP000002282 UP000075903 UP000075880 UP000008792 UP000008711 UP000075900 UP000001819 UP000000673 UP000075885 UP000192221 UP000007266 UP000069272 UP000075883 UP000075920 UP000076502 UP000076408 UP000069940 UP000249989 UP000092553 UP000007798 UP000078541 UP000075882 UP000078492 UP000053825 UP000078540 UP000053097 UP000005205 UP000075809
UP000095301 UP000001070 UP000007801 UP000091820 UP000009192 UP000075881 UP000075884 UP000007062 UP000000304 UP000075902 UP000095300 UP000075840 UP000192223 UP000268350 UP000000803 UP000076407 UP000075886 UP000002282 UP000075903 UP000075880 UP000008792 UP000008711 UP000075900 UP000001819 UP000000673 UP000075885 UP000192221 UP000007266 UP000069272 UP000075883 UP000075920 UP000076502 UP000076408 UP000069940 UP000249989 UP000092553 UP000007798 UP000078541 UP000075882 UP000078492 UP000053825 UP000078540 UP000053097 UP000005205 UP000075809
Interpro
ProteinModelPortal
H9JS58
A0A2W1BUF7
A0A2A4K7K6
A0A212F8J7
A0A2J7Q344
A0A0L0BWL3
+ More
A0A067QZ58 W8AQQ8 T1PLE3 T1PCU7 A0A1I8MNN3 W8B4Z9 A0A034V8A2 A0A034V5K0 B4J334 A0A0P8YFC8 A0A0K8WGP6 A0A0P8XSH9 B3M7T3 A0A1A9WEC2 B4L068 A0A182K428 A0A182N5V8 Q8T5I9 B4QR94 A0A0J9RVB7 A0A182TW57 A0A1I8NN16 A0A1I8NN17 A0A182HL76 A0A1W4WT68 A0A3B0JZA7 A0A1S4GCA3 Q9VTW8 A0A182WSC5 A0A3B0K266 A0A182QW38 B4PGC5 A0A1W4WH00 A0A0A9WLJ8 A0A3B0JZD4 A0A182V675 A0A182IUZ5 A0A0K8THF8 A0A0Q9WQ97 A0A0J9RUA0 B3NH95 B4LFG0 A0A182R4U3 Q2M0N5 A0A2M4CSC4 A0A2M3YYV0 A0A0R3P7S7 W5JDN1 A0A0R1DXN2 A0A2M4CSL5 A0A2M4BB00 M9PCA7 A0A182PGT7 A0A1W4UGX5 A0A2M3ZY80 A0A2M4BCB9 D2A527 A0A2M4CSF9 A0A2M3ZYG8 A0A182FBN2 A0A182MPU2 A0A0Q5U446 A0A1W4UU50 A0A182VWK0 A0A154PG76 A0A182YAE0 A0A0Q9WVL2 A0A0J9RU03 A0A1Q3FV52 A0A1Q3FYY0 A0A182GFY2 A0A0R1E2A3 A0A1B6CXS0 A0A1B6D1J5 A0A1B6D5F3 A0A0M4ELI8 B4N3I8 A0A195FZH4 A0A182L9J3 A0A1B6KU44 A0A1Y1L619 A0A195DRW0 A0A0L7R055 A0A195BTY2 A0A1B6I7U9 A0A1B6K8B8 A0A026WBV0 A0A158NVA6 A0A1B6K518 A0A0J9UJU1 M9PI37 A0A0R1DXU5 A0A0Q5UIZ1 A0A151WUM5
A0A067QZ58 W8AQQ8 T1PLE3 T1PCU7 A0A1I8MNN3 W8B4Z9 A0A034V8A2 A0A034V5K0 B4J334 A0A0P8YFC8 A0A0K8WGP6 A0A0P8XSH9 B3M7T3 A0A1A9WEC2 B4L068 A0A182K428 A0A182N5V8 Q8T5I9 B4QR94 A0A0J9RVB7 A0A182TW57 A0A1I8NN16 A0A1I8NN17 A0A182HL76 A0A1W4WT68 A0A3B0JZA7 A0A1S4GCA3 Q9VTW8 A0A182WSC5 A0A3B0K266 A0A182QW38 B4PGC5 A0A1W4WH00 A0A0A9WLJ8 A0A3B0JZD4 A0A182V675 A0A182IUZ5 A0A0K8THF8 A0A0Q9WQ97 A0A0J9RUA0 B3NH95 B4LFG0 A0A182R4U3 Q2M0N5 A0A2M4CSC4 A0A2M3YYV0 A0A0R3P7S7 W5JDN1 A0A0R1DXN2 A0A2M4CSL5 A0A2M4BB00 M9PCA7 A0A182PGT7 A0A1W4UGX5 A0A2M3ZY80 A0A2M4BCB9 D2A527 A0A2M4CSF9 A0A2M3ZYG8 A0A182FBN2 A0A182MPU2 A0A0Q5U446 A0A1W4UU50 A0A182VWK0 A0A154PG76 A0A182YAE0 A0A0Q9WVL2 A0A0J9RU03 A0A1Q3FV52 A0A1Q3FYY0 A0A182GFY2 A0A0R1E2A3 A0A1B6CXS0 A0A1B6D1J5 A0A1B6D5F3 A0A0M4ELI8 B4N3I8 A0A195FZH4 A0A182L9J3 A0A1B6KU44 A0A1Y1L619 A0A195DRW0 A0A0L7R055 A0A195BTY2 A0A1B6I7U9 A0A1B6K8B8 A0A026WBV0 A0A158NVA6 A0A1B6K518 A0A0J9UJU1 M9PI37 A0A0R1DXU5 A0A0Q5UIZ1 A0A151WUM5
Ontologies
GO
GO:0006811
GO:0005215
GO:0016021
GO:0055085
GO:0015377
GO:0008511
GO:0005886
GO:0005623
GO:0015379
GO:0055078
GO:0035725
GO:1902476
GO:0015081
GO:0015378
GO:0055064
GO:0055075
GO:0006884
GO:1990573
GO:0015171
GO:0006813
GO:0003333
GO:0006821
GO:0006810
GO:0016020
GO:0005524
GO:0003909
GO:0051103
GO:0000049
GO:0005975
GO:0004553
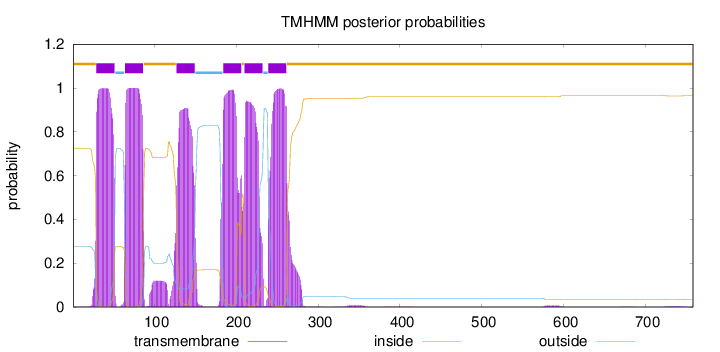
Topology
Length:
758
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
134.68864
Exp number, first 60 AAs:
22.50201
Total prob of N-in:
0.27644
POSSIBLE N-term signal
sequence
outside
1 - 28
TMhelix
29 - 51
inside
52 - 63
TMhelix
64 - 86
outside
87 - 126
TMhelix
127 - 149
inside
150 - 183
TMhelix
184 - 206
outside
207 - 209
TMhelix
210 - 232
inside
233 - 238
TMhelix
239 - 261
outside
262 - 758
Population Genetic Test Statistics
Pi
329.151071
Theta
179.88979
Tajima's D
2.9042
CLR
0.769478
CSRT
0.975301234938253
Interpretation
Uncertain
