Pre Gene Modal
BGIBMGA012371
Annotation
PREDICTED:_zinc_transporter_ZIP3_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.468
Sequence
CDS
ATGAATGTGACCACGGCCAAGGTCGTGTCGATGCTGGCGCTGGGCTTAGGGAGTTTCTTTGGTGGTATGTTGCCTTCATGTTTTTCGGAAAGCGCAAGACAACGGCACCCTCTCGTGCTATCCTGCCTCCTCTGCTTCGGTGGTGGAGTGCTTCTGGCCACTTCCCTCGTCCACATGTTGCCAGAAGCAAGGGAAAAATACCCGAACTACTCAGAGCTAATGGTCTGCATTGGTTTCTTCATTGTTTACTTGGTTGATGAGATATTCCATCTGTGTTCCACTGAAATCAATCATGGAAGTCCGGCCAATCAGGGCTTGAGATATGGAACTGAGAGATCGTCTTTGCTGAATCGCGATGGAGAATTGAGAGAACGATGTCCCGGTGAGGTGGATAACCCTCAAATGTGCCACGTGAGTCATACGGCTCCGTGCAATAGGAGTTCATCAGGAACAATAGGATTGTTATGCGCTCTCTTCGTTCACAGCCTACTGGAAGGTCTGGCTATTGGTCTTCAAGAAGATGGAACTCAGGTGCTGCTTCTGCTAGGAGCTGTCGCCTCTCACAAGTTTGTCGTGGCGTTTTGCCTCGGCGCTGAGCTGTGCGCTAGTCTGCCGGGCCGGATGTGCGCCCAAATGGTGTGCATCCTTATATTCAGCGGTGGCTCCGTCGCCGGAATCGGCATGGGAGCAGCCTTGGAAAATGTCAGAACGCTGAAGGATTCTGTGGCGGTACCGATTATGCAGGCACTTGCGGCGGGAACACTGCTTTACGTGACTGTGAGCGAAGTTCTTCCACGAGAGAGAGCAAATTGGCACGAGCGCAAGAGCACCGCTGGTATTGCACAATTTACAGCAGTAGCAGTCGGCTTCGCGGCTATGTATTTGTGCACTGTATTCCTAGATCATGACGGTTGA
Protein
MNVTTAKVVSMLALGLGSFFGGMLPSCFSESARQRHPLVLSCLLCFGGGVLLATSLVHMLPEAREKYPNYSELMVCIGFFIVYLVDEIFHLCSTEINHGSPANQGLRYGTERSSLLNRDGELRERCPGEVDNPQMCHVSHTAPCNRSSSGTIGLLCALFVHSLLEGLAIGLQEDGTQVLLLLGAVASHKFVVAFCLGAELCASLPGRMCAQMVCILIFSGGSVAGIGMGAALENVRTLKDSVAVPIMQALAAGTLLYVTVSEVLPRERANWHERKSTAGIAQFTAVAVGFAAMYLCTVFLDHDG
Summary
Uniprot
H9JS61
S4PGZ1
A0A3S2NUY3
A0A2W1BSI2
I4DQ93
A0A194RB24
+ More
A0A1E1WH21 A0A194PWD6 A0A2A4K6U3 A0A212F8L9 A0A1L8DED8 A0A1L8DED1 A0A1L8DE73 A0A336KBR9 A0A336K4L5 A0A336LMG1 A0A1L8DDL2 A0A1I8NCI3 A0A1Y1LZ07 A0A1Y1LXY9 B4NIS1 B4GLB4 A0A1W4VGD4 Q294B5 B3MX86 A0A3B0KJJ6 B4LZZ0 A0A3B0JNW8 A0A1I8Q4B9 A0A0L0CPI4 B4K5V1 A0A1J1IE06 A0A087ZVP2 A0A0C9R835 W8BEG7 A0A2A3E864 A0A1A9W721 A0A182QJZ9 A0A182KDN8 A0A310SCT2 A0A0L7RFY5 Q16I91 B4PS63 E2BBB4 A0A0R1E3H4 Q9VFA2 B4HDZ1 A0A023ERK3 A0A1I8P9G5 A0A154PFF8 A0A084W6L1 B3P0T4 A0A1I8JW76 A0A182UAK1 A0A2C9GR55 A0A182V444 Q7QCH1 A0A151I096 B4JGC8 A0A182SMA2 A0A182MIX3 A0A034VV61 A0A026WVL2 A0A1S4FAQ0 A0A3L8D4G3 Q179W9 A0A0K8V9N2 A0A182IVW3 B4QZ24 A0A182PI61 A0A182Y6L3 A0A023EN88 A0A151IDK6 A0A182RXQ5 A0A1I8PPN0 A0A0M9A5F6 A0A1Q3FA34 A0A1Q3FA84 B0WGZ0 A0A2M3ZKU6 A0A2M4BST0 A0A151JXA3 A0A2M4BSS1 A0A2M4BSS7 A0A182GXA1 A0A1B6HYB8 E2A8S9 A0A151WS37 F4X450 A0A195EE58 A0A1B6KG26 B0WZN0 W5JAW3 A0A0K8WCA7 A0A0K8TLB6 A0A182W0T4 A0A023EMU7
A0A1E1WH21 A0A194PWD6 A0A2A4K6U3 A0A212F8L9 A0A1L8DED8 A0A1L8DED1 A0A1L8DE73 A0A336KBR9 A0A336K4L5 A0A336LMG1 A0A1L8DDL2 A0A1I8NCI3 A0A1Y1LZ07 A0A1Y1LXY9 B4NIS1 B4GLB4 A0A1W4VGD4 Q294B5 B3MX86 A0A3B0KJJ6 B4LZZ0 A0A3B0JNW8 A0A1I8Q4B9 A0A0L0CPI4 B4K5V1 A0A1J1IE06 A0A087ZVP2 A0A0C9R835 W8BEG7 A0A2A3E864 A0A1A9W721 A0A182QJZ9 A0A182KDN8 A0A310SCT2 A0A0L7RFY5 Q16I91 B4PS63 E2BBB4 A0A0R1E3H4 Q9VFA2 B4HDZ1 A0A023ERK3 A0A1I8P9G5 A0A154PFF8 A0A084W6L1 B3P0T4 A0A1I8JW76 A0A182UAK1 A0A2C9GR55 A0A182V444 Q7QCH1 A0A151I096 B4JGC8 A0A182SMA2 A0A182MIX3 A0A034VV61 A0A026WVL2 A0A1S4FAQ0 A0A3L8D4G3 Q179W9 A0A0K8V9N2 A0A182IVW3 B4QZ24 A0A182PI61 A0A182Y6L3 A0A023EN88 A0A151IDK6 A0A182RXQ5 A0A1I8PPN0 A0A0M9A5F6 A0A1Q3FA34 A0A1Q3FA84 B0WGZ0 A0A2M3ZKU6 A0A2M4BST0 A0A151JXA3 A0A2M4BSS1 A0A2M4BSS7 A0A182GXA1 A0A1B6HYB8 E2A8S9 A0A151WS37 F4X450 A0A195EE58 A0A1B6KG26 B0WZN0 W5JAW3 A0A0K8WCA7 A0A0K8TLB6 A0A182W0T4 A0A023EMU7
Pubmed
19121390
23622113
28756777
22651552
26354079
22118469
+ More
25315136 28004739 17994087 15632085 26108605 24495485 17510324 17550304 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24945155 24438588 12364791 14747013 17210077 25348373 24508170 30249741 25244985 26483478 21719571 20920257 23761445 26369729
25315136 28004739 17994087 15632085 26108605 24495485 17510324 17550304 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24945155 24438588 12364791 14747013 17210077 25348373 24508170 30249741 25244985 26483478 21719571 20920257 23761445 26369729
EMBL
BABH01033144
BABH01033145
GAIX01003452
JAA89108.1
RSAL01000066
RVE49374.1
+ More
KZ149932 PZC77291.1 AK403995 BAM20083.1 KQ460473 KPJ14480.1 GDQN01004750 JAT86304.1 KQ459591 KPI97069.1 NWSH01000093 PCG79638.1 AGBW02009707 OWR50068.1 GFDF01009337 JAV04747.1 GFDF01009339 JAV04745.1 GFDF01009338 JAV04746.1 UFQS01000046 UFQT01000046 SSW98520.1 SSX18906.1 SSW98518.1 SSX18904.1 SSW98519.1 SSX18905.1 GFDF01009556 JAV04528.1 GEZM01045059 JAV77948.1 GEZM01045058 GEZM01045056 JAV77951.1 CH964272 EDW83785.1 CH479185 EDW38338.1 CM000070 EAL29049.3 CH902629 EDV35309.1 OUUW01000008 SPP83928.1 CH940650 EDW67218.1 SPP83927.1 JRES01000091 KNC34171.1 CH933806 EDW15163.1 CVRI01000047 CRK97788.1 GBYB01009037 JAG78804.1 GAMC01006870 JAB99685.1 KZ288327 PBC27953.1 AXCN02001464 KQ769895 OAD52776.1 KQ414599 KOC69748.1 CH478095 EAT33987.1 CM000160 EDW97483.1 GL446968 EFN87022.1 KRK03773.1 AE014297 BT150206 AAF55158.2 AGT97038.1 CH480815 EDW42083.1 GAPW01001768 JAC11830.1 KQ434893 KZC10593.1 ATLV01020841 KE525308 KFB45855.1 CH954181 EDV48982.1 APCN01000278 AAAB01008859 EAA08092.5 EGK96833.1 KQ976687 KYM78132.1 CH916369 EDV92597.1 AXCM01000472 GAKP01013514 JAC45438.1 KK107083 EZA60090.1 QOIP01000014 RLU15114.1 CH477344 EAT43029.1 GDHF01016781 JAI35533.1 CM000364 EDX12852.1 GAPW01002741 JAC10857.1 KQ977939 KYM98530.1 KQ435750 KOX76239.1 GFDL01010600 JAV24445.1 GFDL01010550 JAV24495.1 DS231931 EDS27315.1 GGFM01008413 MBW29164.1 GGFJ01006996 MBW56137.1 KQ981607 KYN39400.1 GGFJ01006995 MBW56136.1 GGFJ01006994 MBW56135.1 JXUM01095023 JXUM01095024 KQ564167 KXJ72646.1 GECU01028030 JAS79676.1 GL437663 EFN70158.1 KQ982796 KYQ50607.1 GL888625 EGI58774.1 KQ979074 KYN23087.1 GEBQ01029560 JAT10417.1 DS232211 EDS37544.1 ADMH02002014 ETN59985.1 GDHF01003620 JAI48694.1 GDAI01002647 JAI14956.1 GAPW01002905 JAC10693.1
KZ149932 PZC77291.1 AK403995 BAM20083.1 KQ460473 KPJ14480.1 GDQN01004750 JAT86304.1 KQ459591 KPI97069.1 NWSH01000093 PCG79638.1 AGBW02009707 OWR50068.1 GFDF01009337 JAV04747.1 GFDF01009339 JAV04745.1 GFDF01009338 JAV04746.1 UFQS01000046 UFQT01000046 SSW98520.1 SSX18906.1 SSW98518.1 SSX18904.1 SSW98519.1 SSX18905.1 GFDF01009556 JAV04528.1 GEZM01045059 JAV77948.1 GEZM01045058 GEZM01045056 JAV77951.1 CH964272 EDW83785.1 CH479185 EDW38338.1 CM000070 EAL29049.3 CH902629 EDV35309.1 OUUW01000008 SPP83928.1 CH940650 EDW67218.1 SPP83927.1 JRES01000091 KNC34171.1 CH933806 EDW15163.1 CVRI01000047 CRK97788.1 GBYB01009037 JAG78804.1 GAMC01006870 JAB99685.1 KZ288327 PBC27953.1 AXCN02001464 KQ769895 OAD52776.1 KQ414599 KOC69748.1 CH478095 EAT33987.1 CM000160 EDW97483.1 GL446968 EFN87022.1 KRK03773.1 AE014297 BT150206 AAF55158.2 AGT97038.1 CH480815 EDW42083.1 GAPW01001768 JAC11830.1 KQ434893 KZC10593.1 ATLV01020841 KE525308 KFB45855.1 CH954181 EDV48982.1 APCN01000278 AAAB01008859 EAA08092.5 EGK96833.1 KQ976687 KYM78132.1 CH916369 EDV92597.1 AXCM01000472 GAKP01013514 JAC45438.1 KK107083 EZA60090.1 QOIP01000014 RLU15114.1 CH477344 EAT43029.1 GDHF01016781 JAI35533.1 CM000364 EDX12852.1 GAPW01002741 JAC10857.1 KQ977939 KYM98530.1 KQ435750 KOX76239.1 GFDL01010600 JAV24445.1 GFDL01010550 JAV24495.1 DS231931 EDS27315.1 GGFM01008413 MBW29164.1 GGFJ01006996 MBW56137.1 KQ981607 KYN39400.1 GGFJ01006995 MBW56136.1 GGFJ01006994 MBW56135.1 JXUM01095023 JXUM01095024 KQ564167 KXJ72646.1 GECU01028030 JAS79676.1 GL437663 EFN70158.1 KQ982796 KYQ50607.1 GL888625 EGI58774.1 KQ979074 KYN23087.1 GEBQ01029560 JAT10417.1 DS232211 EDS37544.1 ADMH02002014 ETN59985.1 GDHF01003620 JAI48694.1 GDAI01002647 JAI14956.1 GAPW01002905 JAC10693.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000218220
UP000007151
+ More
UP000095301 UP000007798 UP000008744 UP000192221 UP000001819 UP000007801 UP000268350 UP000008792 UP000095300 UP000037069 UP000009192 UP000183832 UP000005203 UP000242457 UP000091820 UP000075886 UP000075881 UP000053825 UP000008820 UP000002282 UP000008237 UP000000803 UP000001292 UP000076502 UP000030765 UP000008711 UP000076407 UP000075902 UP000075840 UP000075903 UP000007062 UP000078540 UP000001070 UP000075901 UP000075883 UP000053097 UP000279307 UP000075880 UP000000304 UP000075885 UP000076408 UP000078542 UP000075900 UP000053105 UP000002320 UP000078541 UP000069940 UP000249989 UP000000311 UP000075809 UP000007755 UP000078492 UP000000673 UP000075920
UP000095301 UP000007798 UP000008744 UP000192221 UP000001819 UP000007801 UP000268350 UP000008792 UP000095300 UP000037069 UP000009192 UP000183832 UP000005203 UP000242457 UP000091820 UP000075886 UP000075881 UP000053825 UP000008820 UP000002282 UP000008237 UP000000803 UP000001292 UP000076502 UP000030765 UP000008711 UP000076407 UP000075902 UP000075840 UP000075903 UP000007062 UP000078540 UP000001070 UP000075901 UP000075883 UP000053097 UP000279307 UP000075880 UP000000304 UP000075885 UP000076408 UP000078542 UP000075900 UP000053105 UP000002320 UP000078541 UP000069940 UP000249989 UP000000311 UP000075809 UP000007755 UP000078492 UP000000673 UP000075920
Interpro
Gene 3D
ProteinModelPortal
H9JS61
S4PGZ1
A0A3S2NUY3
A0A2W1BSI2
I4DQ93
A0A194RB24
+ More
A0A1E1WH21 A0A194PWD6 A0A2A4K6U3 A0A212F8L9 A0A1L8DED8 A0A1L8DED1 A0A1L8DE73 A0A336KBR9 A0A336K4L5 A0A336LMG1 A0A1L8DDL2 A0A1I8NCI3 A0A1Y1LZ07 A0A1Y1LXY9 B4NIS1 B4GLB4 A0A1W4VGD4 Q294B5 B3MX86 A0A3B0KJJ6 B4LZZ0 A0A3B0JNW8 A0A1I8Q4B9 A0A0L0CPI4 B4K5V1 A0A1J1IE06 A0A087ZVP2 A0A0C9R835 W8BEG7 A0A2A3E864 A0A1A9W721 A0A182QJZ9 A0A182KDN8 A0A310SCT2 A0A0L7RFY5 Q16I91 B4PS63 E2BBB4 A0A0R1E3H4 Q9VFA2 B4HDZ1 A0A023ERK3 A0A1I8P9G5 A0A154PFF8 A0A084W6L1 B3P0T4 A0A1I8JW76 A0A182UAK1 A0A2C9GR55 A0A182V444 Q7QCH1 A0A151I096 B4JGC8 A0A182SMA2 A0A182MIX3 A0A034VV61 A0A026WVL2 A0A1S4FAQ0 A0A3L8D4G3 Q179W9 A0A0K8V9N2 A0A182IVW3 B4QZ24 A0A182PI61 A0A182Y6L3 A0A023EN88 A0A151IDK6 A0A182RXQ5 A0A1I8PPN0 A0A0M9A5F6 A0A1Q3FA34 A0A1Q3FA84 B0WGZ0 A0A2M3ZKU6 A0A2M4BST0 A0A151JXA3 A0A2M4BSS1 A0A2M4BSS7 A0A182GXA1 A0A1B6HYB8 E2A8S9 A0A151WS37 F4X450 A0A195EE58 A0A1B6KG26 B0WZN0 W5JAW3 A0A0K8WCA7 A0A0K8TLB6 A0A182W0T4 A0A023EMU7
A0A1E1WH21 A0A194PWD6 A0A2A4K6U3 A0A212F8L9 A0A1L8DED8 A0A1L8DED1 A0A1L8DE73 A0A336KBR9 A0A336K4L5 A0A336LMG1 A0A1L8DDL2 A0A1I8NCI3 A0A1Y1LZ07 A0A1Y1LXY9 B4NIS1 B4GLB4 A0A1W4VGD4 Q294B5 B3MX86 A0A3B0KJJ6 B4LZZ0 A0A3B0JNW8 A0A1I8Q4B9 A0A0L0CPI4 B4K5V1 A0A1J1IE06 A0A087ZVP2 A0A0C9R835 W8BEG7 A0A2A3E864 A0A1A9W721 A0A182QJZ9 A0A182KDN8 A0A310SCT2 A0A0L7RFY5 Q16I91 B4PS63 E2BBB4 A0A0R1E3H4 Q9VFA2 B4HDZ1 A0A023ERK3 A0A1I8P9G5 A0A154PFF8 A0A084W6L1 B3P0T4 A0A1I8JW76 A0A182UAK1 A0A2C9GR55 A0A182V444 Q7QCH1 A0A151I096 B4JGC8 A0A182SMA2 A0A182MIX3 A0A034VV61 A0A026WVL2 A0A1S4FAQ0 A0A3L8D4G3 Q179W9 A0A0K8V9N2 A0A182IVW3 B4QZ24 A0A182PI61 A0A182Y6L3 A0A023EN88 A0A151IDK6 A0A182RXQ5 A0A1I8PPN0 A0A0M9A5F6 A0A1Q3FA34 A0A1Q3FA84 B0WGZ0 A0A2M3ZKU6 A0A2M4BST0 A0A151JXA3 A0A2M4BSS1 A0A2M4BSS7 A0A182GXA1 A0A1B6HYB8 E2A8S9 A0A151WS37 F4X450 A0A195EE58 A0A1B6KG26 B0WZN0 W5JAW3 A0A0K8WCA7 A0A0K8TLB6 A0A182W0T4 A0A023EMU7
PDB
5TSB
E-value=0.00240002,
Score=95
Ontologies
GO
Topology
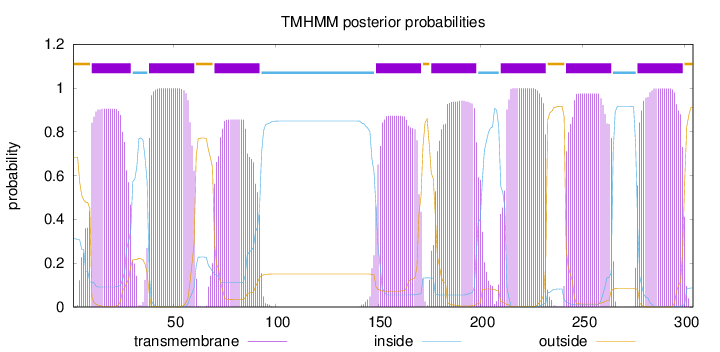
Length:
304
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
163.85344
Exp number, first 60 AAs:
41.18462
Total prob of N-in:
0.31566
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 29
inside
30 - 37
TMhelix
38 - 60
outside
61 - 69
TMhelix
70 - 92
inside
93 - 148
TMhelix
149 - 171
outside
172 - 175
TMhelix
176 - 198
inside
199 - 209
TMhelix
210 - 232
outside
233 - 241
TMhelix
242 - 264
inside
265 - 276
TMhelix
277 - 299
outside
300 - 304
Population Genetic Test Statistics
Pi
154.611903
Theta
147.767006
Tajima's D
0
CLR
0.097568
CSRT
0.367081645917704
Interpretation
Uncertain
