Gene
KWMTBOMO12363
Pre Gene Modal
BGIBMGA012372
Annotation
PREDICTED:_uncharacterized_protein_LOC103572275_[Microplitis_demolitor]
Full name
ATP-dependent DNA helicase
Location in the cell
Nuclear Reliability : 1.815 PlasmaMembrane Reliability : 1.657
Sequence
CDS
ATGCCAAGAAAACGCAAAGGGGCTGATTTGAGCCGCAGTACAAGTAAAGCTCGAAACTTGCGAAATAGCAGATCCGAAAGAACAGAAGAACAAATCCAGCAACAAAATACTGATGCACGTGTTAGAATGGCGCAATTGCATCAAGAAGAGCTAGAAGATACACGAGCTGAACGCAATGAAGTCAGAAGATTAGAACAACGACAATCACGCCGTTTCACAGTCAATAGACGAAGAACAAATGACCAACAACGACAACAGGTACATCGAGCATTTATATCTGATTCATTCCTGCGTCTAGCATTCCAGTATGAGCCCGATATTGAATATCATGCTCATTCAAAAGTGGTAATTGGTGCTATGGACAAGGAATGTCCGCATTGTCATGCTCTGAAATTCAAAAATGAGCCAGCTGGGATGTGTTGCGCGTCAGGAAAAGTGCAACTACCTGAAATTGAAACACCACCTGAACCATTGAACGGCTTACTTATCGGCACGGATCCAGATTCTAACGTGTTCCTGAAGTCAATTCGAAGATTCAATTCATGCTTTCAAATGACATCGTTCGGAGCAACAGAAATAGTTCGAAATACTAATGCAAATGGTCAACAATTCAATTCTACATTCAAAATCAGAGGCCAAGTTTATCATAAAATTGGCTCACTGCTGCCAATGCCAAACGAACCACATGAATTCTTACAAATCTACTTTATGGGCGGCGAGGATTCCGGAAGCGCACTTGCCAATCGCGTGAATGCACGTTGTGATTATAATAACCTTGATTCACTTTATGCCAGGCGCATCGTCAGCGAGCTAGATGCTCTTTTGAACGAGCACAACGAGTTGTTGAATATATTCAAATCACATATGCACCAATTACAAAGCGATAATCACGCTATCGTCATTAATCCTGATAAAACACCAGCTGGAGAGCATATTCGTAGATTCAATGCACCCGTTGTTGATGATGTTGCTGGAATCATGGTTGGCGATTGTACAGCTGCACGAGAAATTGTGATTCGTAGAAGAAATAATAATCTTCAGTTCATTGCTGACACACATCGTTTATATGACGCTCTCCAATATCCGCTAATATTCTGGAAGGGACAAGACGGATATTGCATAAACATAAAACAACGAGATCCCGTATCAGGAGCTGAAACAAACAAGAACGTTAGCTCAAAGGATTATTATGCGTACCGATTAATGATTAGACGTGGACTGGACAACGTCATTTTACGATGTCGTGAGCTTTGTCAACAATTCATGGTCGACATGTACGCGAAGATTGAGAGCGAACGACTACGATATTTACGATACAATCAACAAAAGCTGCGCGCGGAAGAGTACATTCATTTGCGAGACGCTATCAACAACAACGCCGACGTCGCCGAAATTGGTAACCATGTCATTTTACCATCATCGTACGTAGGCAGTCCACGTCATATGCAAGAATATATACAGGATGCTCTGACTTTCGTGCGCGAATATGGACGACCATGTTTATTTATCACGTTCACATGTAATCCAAAATGGCCAGAGATTACATCTTTGCTACTGCCTGGCCAAAATGCAATACATCGCCATGACATTACAGCACGTGTGTTCAGACAAAAGTTGAAGTCTTTAATAAGTTTCATTACTAAATCACATGTATTTGGTCCCACACGTTGCTGGATGTATTCGGTTGAGTGGCAAAAGCGAGGATTACCTCATGCACACATTTTGGTTTGGTTCATCGACAAAATCCGTCCTGAAGAAATCGATAGTATCATTTCTGCGGAAATTCCAGATCCATCCACTGACCAACTGCTGTTTGATGTTGTTACAACAAACATGATTCATGGTCCATGTGTGAATAAAAACGATGAAATAACGCTCTACCAAATTGGTCGGTACATCAGCTCTAATGAAGCTGCTTGGCGTATCTTTGGTTTTCCAATTCATGAACGGGATCCAGCAGTTATTCAGTTAGCCATCCATCTTGAAAACGGTCAGCGTGTATTTTTCACGAACGAGACAGCGATTGATCGTGCAATAAATCCACCTAAAACTACACTCACTGCATTTTTTGAATTGTGTAATCGTGCGGATGAATTTGGTGCCTTTGCACGAACATTACTCTATTCACAAGTACCACGCTATTTCACATGGACTCAAACAAAAACATGGATGCCCCGCAAGCAAGGCTCACCAGTTGCTGCATGTCTCAATTTATTTAATTCAAACGCCTTGGGGCGATTATTTACAGTCAATCCAAGACACACGGAGTGCTTTTATCTTCGACTGTTGTTGGTTAATGTTACTGGCCCATTATCATTTCAAGATATACGTAAAGTGAATGGGCAACAATATCCAACGTATAAAGATGCATGTCTTGCACTCGGCTTGCTGGAAGACGACAACCAGTGGGAATGCATGCTTGCTGAAGCTGCATTGAACTGTACAGCAATACAAATTCGTCTACTATTCGCTATAGTGTTGACTACATGTTTCCCAGCCCGAGCACAGATATTATGGGAAAATCACAAAGATTCAATGACTGATGATATATTGCATCAACACCGTATACGGTGCCACGATCTAACCATAACATTCAGCGACGAAATGTACAATGAAGCATTGATTGCTATTGAGGATCTTTGCATTGTCATTGCCAACTTACCACTTAGTAATTTCGGTATGAATTCGCCAAATCGAACTGCATCTGATTTAATGAATACTGAAATGAATCGTGAACTGCAGTACAGTACTGTAGAAATGGCAGCGATTGTTGCCCGCAATGTCCCACTAATGAATGAGGAACAAAGAACCATTTATGATCGCATTATGCTCGCAGTTTCAGCTGGACAAGGTGGGTTCTTCTTTTTGGATGCACCGGGTGGAACTGGCAAAACATTCGTTATTTCGCTAATTCTTGCTGAAATACGATCAAATAATGGCATCGCATTGGCCGTTGCATCATCGGGCATTGCAGCAACTTTATTGGATGGAGAATAA
Protein
MPRKRKGADLSRSTSKARNLRNSRSERTEEQIQQQNTDARVRMAQLHQEELEDTRAERNEVRRLEQRQSRRFTVNRRRTNDQQRQQVHRAFISDSFLRLAFQYEPDIEYHAHSKVVIGAMDKECPHCHALKFKNEPAGMCCASGKVQLPEIETPPEPLNGLLIGTDPDSNVFLKSIRRFNSCFQMTSFGATEIVRNTNANGQQFNSTFKIRGQVYHKIGSLLPMPNEPHEFLQIYFMGGEDSGSALANRVNARCDYNNLDSLYARRIVSELDALLNEHNELLNIFKSHMHQLQSDNHAIVINPDKTPAGEHIRRFNAPVVDDVAGIMVGDCTAAREIVIRRRNNNLQFIADTHRLYDALQYPLIFWKGQDGYCINIKQRDPVSGAETNKNVSSKDYYAYRLMIRRGLDNVILRCRELCQQFMVDMYAKIESERLRYLRYNQQKLRAEEYIHLRDAINNNADVAEIGNHVILPSSYVGSPRHMQEYIQDALTFVREYGRPCLFITFTCNPKWPEITSLLLPGQNAIHRHDITARVFRQKLKSLISFITKSHVFGPTRCWMYSVEWQKRGLPHAHILVWFIDKIRPEEIDSIISAEIPDPSTDQLLFDVVTTNMIHGPCVNKNDEITLYQIGRYISSNEAAWRIFGFPIHERDPAVIQLAIHLENGQRVFFTNETAIDRAINPPKTTLTAFFELCNRADEFGAFARTLLYSQVPRYFTWTQTKTWMPRKQGSPVAACLNLFNSNALGRLFTVNPRHTECFYLRLLLVNVTGPLSFQDIRKVNGQQYPTYKDACLALGLLEDDNQWECMLAEAALNCTAIQIRLLFAIVLTTCFPARAQILWENHKDSMTDDILHQHRIRCHDLTITFSDEMYNEALIAIEDLCIVIANLPLSNFGMNSPNRTASDLMNTEMNRELQYSTVEMAAIVARNVPLMNEEQRTIYDRIMLAVSAGQGGFFFLDAPGGTGKTFVISLILAEIRSNNGIALAVASSGIAATLLDGE
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the helicase family.
Uniprot
EC Number
3.6.4.12
EMBL
ABLF02032513
ABLF02032521
RSAL01001639
RVE40746.1
KE125664
EPB67478.1
+ More
JARK01001590 EYB87952.1 RSAL01000340 RVE42412.1 ABLF02012908 ABLF02042833 OIVN01000670 SPC83866.1 OIVN01001709 SPC96851.1 LXQA010000149 MCH79500.1 ASHM01001951 PNY07550.1 JN606071 AFK13834.1 NIVC01000291 PAA86185.1 NIVC01004811 PAA46541.1 JARK01001557 EYB90395.1 JOJR01001364 RCN31185.1 ADAS01008033
JARK01001590 EYB87952.1 RSAL01000340 RVE42412.1 ABLF02012908 ABLF02042833 OIVN01000670 SPC83866.1 OIVN01001709 SPC96851.1 LXQA010000149 MCH79500.1 ASHM01001951 PNY07550.1 JN606071 AFK13834.1 NIVC01000291 PAA86185.1 NIVC01004811 PAA46541.1 JARK01001557 EYB90395.1 JOJR01001364 RCN31185.1 ADAS01008033
Proteomes
Pfam
Interpro
ProteinModelPortal
Ontologies
GO
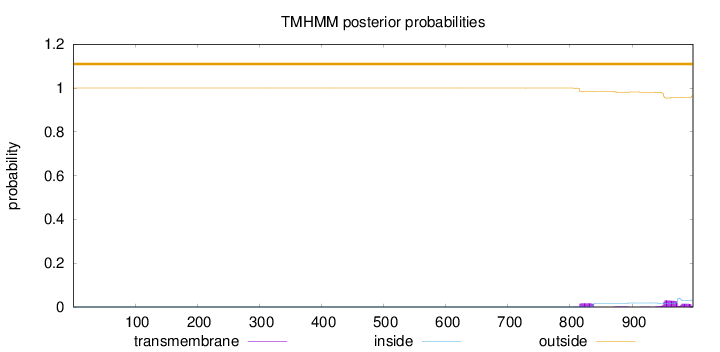
Topology
Length:
998
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.38349
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00006
outside
1 - 998
Population Genetic Test Statistics
Pi
21.330274
Theta
160.124958
Tajima's D
1.825752
CLR
0.051803
CSRT
0.855507224638768
Interpretation
Uncertain
