Gene
KWMTBOMO12362
Pre Gene Modal
BGIBMGA006350
Annotation
PREDICTED:_uncharacterized_protein_LOC105392658_[Plutella_xylostella]
Location in the cell
Extracellular Reliability : 2.79
Sequence
CDS
ATGATAAAAGGTGTTCTGTGCAAAATGATTAATTGTAATTTGGGTGACCTCTCTTGGGAGCAATGTGTCCTGCCAGTACGTTTTGGTGGTATCGGCGTACGTAAAGCATCAGCTGTTGCGTTGCCGGCCTTTCTTGCTTCTGCACGCGCCTCCTGTCTCATAGTCGGTAATGTTCTCAACCCTTGTATGGGTTCCGTAGAGATTGTGGGTGTGTCGGAGGCTGGATCGGCCTGGCAGTCTTTTTGTCAGGCGGATTTTCCTACTTTACCGCATTTGCAGCGTCTTTGGGACACTCCGCTTTGTCAACGGATACAGCAACGGTTACTACAGTCATATGATAACAGCGTGGACATTGCTCGTTTTCGCGCCGTTACCGAAAAGGAATCAAGTTTCTGGCTTCAAGCCTATCCATCGTCCAATACAGGCACGTTGTTGGACAACAACGCTCTCTCGCTAGCTATTGGCTTACGGTTGGGGGCCCATATTAACGTACCACATTTCTGTGTCTGTGGCGAGTACGTCAATGAGCTTGGACATCACGGACTCTCGTGCCAGAAAAGCGCTGGGCGCATTCCAAGACACGCATCTTTAAATGATATGATAAGAAGGGCGCTCACTACAGTCAATGTTCCGGCTGTCTTGGAACCCATCGGCATTTTGAGGTCAGATGGGAAAAGGCCCGATGGAGTCACGCTGATTCCATGTGAAAAAGGTTTTTAG
Protein
MIKGVLCKMINCNLGDLSWEQCVLPVRFGGIGVRKASAVALPAFLASARASCLIVGNVLNPCMGSVEIVGVSEAGSAWQSFCQADFPTLPHLQRLWDTPLCQRIQQRLLQSYDNSVDIARFRAVTEKESSFWLQAYPSSNTGTLLDNNALSLAIGLRLGAHINVPHFCVCGEYVNELGHHGLSCQKSAGRIPRHASLNDMIRRALTTVNVPAVLEPIGILRSDGKRPDGVTLIPCEKGF
Summary
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Uniprot
A0A194Q0M9
H9ISD2
A0A1D1V0N9
A0A1D1V1G4
A0A437B3N6
A0A1D1VI15
+ More
A0A1D1V5S7 A0A1D1WA43 A0A1D1W783 A0A1Y1MW21 A0A226DSQ4 A0A1D1UPD4 A0A1W0WJQ4 A0A226EVZ6 A0A1D1VCB8 A0A2P8XZ03 A0A226ENY2 A0A226D463 A0A1D1VFL0 A0A2B7ZYB8 A0A2B7ZW65 A0A226DU26 A0A226DXH2 A0A2V3IC37 A0A2B7ZRL5 A0A1Y1MJ73 A0A1X7U8Q8 A0A1W0X5Z2 A0A1D1VIH0 A0A1D1UV52 T1F592 A0A1X7UWM7 A0A1D1VIZ6 T1FE88 T1ETD9 T1EQU9 T1FL49 A0A1X7UJV4 T1FE76 T1ER08 T1FKY4 T1ESP3 A0A2P8XV61 A0A226F2H2 T1FER8 A0A1X7TVW8 A0A2P8YGC7 A0A1D1W7W2
A0A1D1V5S7 A0A1D1WA43 A0A1D1W783 A0A1Y1MW21 A0A226DSQ4 A0A1D1UPD4 A0A1W0WJQ4 A0A226EVZ6 A0A1D1VCB8 A0A2P8XZ03 A0A226ENY2 A0A226D463 A0A1D1VFL0 A0A2B7ZYB8 A0A2B7ZW65 A0A226DU26 A0A226DXH2 A0A2V3IC37 A0A2B7ZRL5 A0A1Y1MJ73 A0A1X7U8Q8 A0A1W0X5Z2 A0A1D1VIH0 A0A1D1UV52 T1F592 A0A1X7UWM7 A0A1D1VIZ6 T1FE88 T1ETD9 T1EQU9 T1FL49 A0A1X7UJV4 T1FE76 T1ER08 T1FKY4 T1ESP3 A0A2P8XV61 A0A226F2H2 T1FER8 A0A1X7TVW8 A0A2P8YGC7 A0A1D1W7W2
EMBL
KQ459581
KPI99111.1
BABH01024325
BDGG01000003
GAU95301.1
BDGG01000002
+ More
GAU94690.1 RSAL01000186 RVE44793.1 BDGG01000007 GAV01250.1 BDGG01000004 GAU97086.1 BDGG01000021 GAV09288.1 GAV09252.1 GEZM01019101 JAV89874.1 LNIX01000012 OXA48253.1 GAU91574.1 MTYJ01000088 OQV15451.1 LNIX01000001 OXA61805.1 BDGG01000005 GAU99276.1 PYGN01001141 PSN37238.1 LNIX01000003 OXA58286.1 LNIX01000038 OXA39608.1 BDGG01000006 GAV00437.1 PDHN01000629 PGH38391.1 PDHN01001114 PGH37252.1 LNIX01000011 OXA48508.1 LNIX01000010 OXA49381.1 NBIV01000529 PXF39672.1 PDHN01000428 PGH39064.1 GEZM01032301 JAV84680.1 MTYJ01000015 OQV22929.1 GAV01400.1 GAU91622.1 AMQM01004200 KB096457 ESO04719.1 GAV01570.1 AMQM01006745 KB097510 ESN95674.1 AMQM01001231 KB097143 ESN99390.1 AMQM01000699 KB096324 ESO06725.1 AMQM01009916 KB097151 ESN98374.1 AMQM01006742 ESN95658.1 AMQM01000737 KB096742 ESO01848.1 AMQM01009627 KB096717 ESO02733.1 AMQM01001089 ESN98981.1 PYGN01001298 PSN35898.1 OXA63637.1 AMQM01006863 KB097528 ESN95404.1 PYGN01000621 PSN43234.1 BDGG01000023 GAV09477.1
GAU94690.1 RSAL01000186 RVE44793.1 BDGG01000007 GAV01250.1 BDGG01000004 GAU97086.1 BDGG01000021 GAV09288.1 GAV09252.1 GEZM01019101 JAV89874.1 LNIX01000012 OXA48253.1 GAU91574.1 MTYJ01000088 OQV15451.1 LNIX01000001 OXA61805.1 BDGG01000005 GAU99276.1 PYGN01001141 PSN37238.1 LNIX01000003 OXA58286.1 LNIX01000038 OXA39608.1 BDGG01000006 GAV00437.1 PDHN01000629 PGH38391.1 PDHN01001114 PGH37252.1 LNIX01000011 OXA48508.1 LNIX01000010 OXA49381.1 NBIV01000529 PXF39672.1 PDHN01000428 PGH39064.1 GEZM01032301 JAV84680.1 MTYJ01000015 OQV22929.1 GAV01400.1 GAU91622.1 AMQM01004200 KB096457 ESO04719.1 GAV01570.1 AMQM01006745 KB097510 ESN95674.1 AMQM01001231 KB097143 ESN99390.1 AMQM01000699 KB096324 ESO06725.1 AMQM01009916 KB097151 ESN98374.1 AMQM01006742 ESN95658.1 AMQM01000737 KB096742 ESO01848.1 AMQM01009627 KB096717 ESO02733.1 AMQM01001089 ESN98981.1 PYGN01001298 PSN35898.1 OXA63637.1 AMQM01006863 KB097528 ESN95404.1 PYGN01000621 PSN43234.1 BDGG01000023 GAV09477.1
Proteomes
PRIDE
Interpro
IPR000477
RT_dom
+ More
IPR036388 WH-like_DNA-bd_sf
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR019786 Zinc_finger_PHD-type_CS
IPR003150 DNA-bd_RFX
IPR036390 WH_DNA-bd_sf
IPR008962 PapD-like_sf
IPR013783 Ig-like_fold
IPR001873 ENaC
IPR004875 DDE_SF_endonuclease_dom
IPR020903 ENaC_CS
IPR029063 SAM-dependent_MTases
IPR036388 WH-like_DNA-bd_sf
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR019786 Zinc_finger_PHD-type_CS
IPR003150 DNA-bd_RFX
IPR036390 WH_DNA-bd_sf
IPR008962 PapD-like_sf
IPR013783 Ig-like_fold
IPR001873 ENaC
IPR004875 DDE_SF_endonuclease_dom
IPR020903 ENaC_CS
IPR029063 SAM-dependent_MTases
Gene 3D
ProteinModelPortal
A0A194Q0M9
H9ISD2
A0A1D1V0N9
A0A1D1V1G4
A0A437B3N6
A0A1D1VI15
+ More
A0A1D1V5S7 A0A1D1WA43 A0A1D1W783 A0A1Y1MW21 A0A226DSQ4 A0A1D1UPD4 A0A1W0WJQ4 A0A226EVZ6 A0A1D1VCB8 A0A2P8XZ03 A0A226ENY2 A0A226D463 A0A1D1VFL0 A0A2B7ZYB8 A0A2B7ZW65 A0A226DU26 A0A226DXH2 A0A2V3IC37 A0A2B7ZRL5 A0A1Y1MJ73 A0A1X7U8Q8 A0A1W0X5Z2 A0A1D1VIH0 A0A1D1UV52 T1F592 A0A1X7UWM7 A0A1D1VIZ6 T1FE88 T1ETD9 T1EQU9 T1FL49 A0A1X7UJV4 T1FE76 T1ER08 T1FKY4 T1ESP3 A0A2P8XV61 A0A226F2H2 T1FER8 A0A1X7TVW8 A0A2P8YGC7 A0A1D1W7W2
A0A1D1V5S7 A0A1D1WA43 A0A1D1W783 A0A1Y1MW21 A0A226DSQ4 A0A1D1UPD4 A0A1W0WJQ4 A0A226EVZ6 A0A1D1VCB8 A0A2P8XZ03 A0A226ENY2 A0A226D463 A0A1D1VFL0 A0A2B7ZYB8 A0A2B7ZW65 A0A226DU26 A0A226DXH2 A0A2V3IC37 A0A2B7ZRL5 A0A1Y1MJ73 A0A1X7U8Q8 A0A1W0X5Z2 A0A1D1VIH0 A0A1D1UV52 T1F592 A0A1X7UWM7 A0A1D1VIZ6 T1FE88 T1ETD9 T1EQU9 T1FL49 A0A1X7UJV4 T1FE76 T1ER08 T1FKY4 T1ESP3 A0A2P8XV61 A0A226F2H2 T1FER8 A0A1X7TVW8 A0A2P8YGC7 A0A1D1W7W2
Ontologies
KEGG
PANTHER
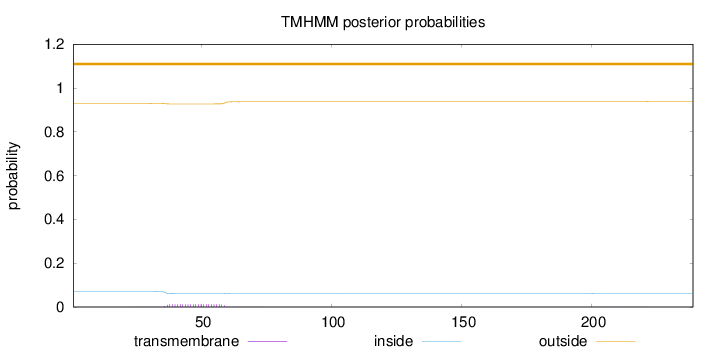
Topology
Length:
239
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.306430000000001
Exp number, first 60 AAs:
0.28813
Total prob of N-in:
0.07087
outside
1 - 239
Population Genetic Test Statistics
Pi
59.113264
Theta
56.595019
Tajima's D
0.376246
CLR
0
CSRT
0.48077596120194
Interpretation
Uncertain
