Gene
KWMTBOMO12361
Pre Gene Modal
BGIBMGA012374
Annotation
PREDICTED:_neither_inactivation_nor_afterpotential_protein_G_[Amyelois_transitella]
Full name
Neither inactivation nor afterpotential protein G
Location in the cell
Mitochondrial Reliability : 1.969
Sequence
CDS
ATGACAATATATACGTACGTCTTAGGTGTGGTTTTCGCATGTTTGTCTTACGTTTTATATCAGAGATATGCGAACGTGTATTCCAGTATTATTAAGAAGCCAGAAAAGAGATATGATTATATTATTGTTGGAGCCGGCACATCAGGTTGCGTCCTGGCCTCCCGCCTCTCCGAAGACCCGAACACTAAAGTCCTTCTAATCGAAGCTGGTGAGCAAATGGGCTACTTCACCAGGATGCCTCTAACGCCGACTGCGGCTCAGCAGGGACCGAACGACTGGTCCGTGAGAACAGTCCCACAAAGATACTCATCTTTTGGACTTTGGCGGCAGACACAAATCCTGCCACGCGGAAAAGGACTCGGCGGCTCCGGACAAATAAACTTCCTCCTTCACGGATCTGGTTCGCCCCGAGATTACGAGAAATGGTCTCAACTCGGCTTCGAAGGTTGGACGTTTGAGGACTTGAAGCCATACTTCTTGAAGGCATTTGGGACAACGGAGAGTGAATTTGACACTGAACCCTGTTACGAAGATGGCTTTTGTCCAAAATCAATGGCGCCAATGAAACTAGTACAAAATCACCGCGACGAACTAATGACGTCATTTAAAGAAGCATCCAGACAGACGAAGACAAAGTCCACAAAGTTTCAAAAAGCCACAGCGACAATTAAGGACGGTGTCAGACACGTCTCGTTCGACGCATACCTGAAGCCGGCACTGCAGAGAGACAACTTACACGTTTTGCTGAACACCCAGGGTATTTCAATACGCTTCGAAAACAAGAAGGCAGTGTCTCTGTACATTTTGCTAAACCATCGATACCTTAACAACGTTTTCATCAACAAGGACATCATTATTTGCGCGGGCGCCATTAAGACACCGCAGATACTAATGCTGTCTGGAATCGGACCGAAAGATTTGTTGCAGAAATTGAAAATAAAGCCGGTTGTGGTGAACGAGCACGTTGGCAGGAACTTGCACGATCATATGAATTTTCCCCTCTACGTCAGCATCCAGAAGCCAATCAGCGTAACCCTGGCGAAGGTATTCACCGCGACGACCGTCTGGGAATACTTGTGGAAAGGCGAAGGTCTACTGTCTTTCCCACCTGTCGCTGGAGTCGAATACCAGAACCAGTCTGCCGTGATGCTGTTCTCCATGGGAACATCTAGTGAGAGACTGCTGCGGGATCTGTCCAATTACAAACCTCAAGTCTTCCGCGCAACCTTTCCATTTCACAACTCGACAAGAAAGGAAGGGTTCATGTTCCTATCGACTTGCACCCAGCCCATCAGCCGAGGGTCTGTCAGCTTAGCGGATGCCAACACGAAAAGTCCCCCCCTCGTAGACCCTCGGTACTTGAAGAGATTCGAGGATGTTAGATGCATGATACAAGCAATAAGGAGGGCAGAGCAGCTGGTGTCAACAAAGCCGTTTCAGGACGTGGGAGCAAAGATCCACTGGCCCAGACCTGAGCGTTGTCTTACCTTCTGGAACTACACGTCGGAAGATCAGAGGAGCAGAAAGCGATTGAGATTACAGCAAAAGCAACAGAAATTTGAAAGAAAGATAAAATCTGCAAAAATCAATACTCCCCCGGATACGTATCTGGAGTGTCTGATACGAGAGGTCGCGGTGACCGGTCACCACGTCGCCGGGACCTGCTCCGGTGGAACCGTCGTTGATGACCAATTACAATTAAAATCAGTTTCCGGCGTTCGCATAATGGACGCCAGCGTACTTCCGGCGCCGTTGTCTCTGTACCCGAACTCTGTGCTAATAGCTATGGCCGAAAAACTCAGTGAAATGATGCGAAATAATATTTGA
Protein
MTIYTYVLGVVFACLSYVLYQRYANVYSSIIKKPEKRYDYIIVGAGTSGCVLASRLSEDPNTKVLLIEAGEQMGYFTRMPLTPTAAQQGPNDWSVRTVPQRYSSFGLWRQTQILPRGKGLGGSGQINFLLHGSGSPRDYEKWSQLGFEGWTFEDLKPYFLKAFGTTESEFDTEPCYEDGFCPKSMAPMKLVQNHRDELMTSFKEASRQTKTKSTKFQKATATIKDGVRHVSFDAYLKPALQRDNLHVLLNTQGISIRFENKKAVSLYILLNHRYLNNVFINKDIIICAGAIKTPQILMLSGIGPKDLLQKLKIKPVVVNEHVGRNLHDHMNFPLYVSIQKPISVTLAKVFTATTVWEYLWKGEGLLSFPPVAGVEYQNQSAVMLFSMGTSSERLLRDLSNYKPQVFRATFPFHNSTRKEGFMFLSTCTQPISRGSVSLADANTKSPPLVDPRYLKRFEDVRCMIQAIRRAEQLVSTKPFQDVGAKIHWPRPERCLTFWNYTSEDQRSRKRLRLQQKQQKFERKIKSAKINTPPDTYLECLIREVAVTGHHVAGTCSGGTVVDDQLQLKSVSGVRIMDASVLPAPLSLYPNSVLIAMAEKLSEMMRNNI
Summary
Description
Oxidoreductase involved in biosynthesis of 3-hydroxyretinal, a chromophore for rhodopsin Rh1. Not responsible for the initial hydroxylation of the retinal ring but rather acts in a subsequent step in chromophore production. May catalyze the conversion of (3R)-3-hydroxyretinol to the 3S enantiomer.
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Keywords
Complete proteome
FAD
Flavoprotein
Glycoprotein
Oxidoreductase
Reference proteome
Secreted
Sensory transduction
Signal
Vision
Feature
chain Neither inactivation nor afterpotential protein G
Uniprot
H9JS64
A0A2W1BE30
A0A2A4JB55
A0A3S2N6W1
A0A2H1V6D6
A0A194R9G6
+ More
A0A212F8K0 A0A0L7LGH9 A0A182QPL4 A0A1Q3FSG1 A0A182GG60 A0A067RK60 A0A151WKL4 A0A182VWI7 A0A1Y9H2T1 E2AXY6 A0A232ESQ0 K7IZS1 A0A182R4V8 A0A026WKI5 A0A1S4FWX8 A0A182PGV0 A0A195ECK8 A0A182T7V6 A0A2C9GQL4 W5J6V5 A0A182UMT5 A0A0L7QPU6 Q380J0 F4WQ87 Q16LE2 A0A182L9I2 A0A1I8JVQ5 A0A158NXM3 A0A182FRF5 A0A084W244 A0A182TH75 A0A336LIB5 B0W2B2 A0A0C9QT70 A0A195EUJ9 A0A195AZZ7 A0A1B0CC23 A0A182MCE4 A0A1B0D8F4 B4JTK0 A0A0L0C7B9 A0A034VJ86 A0A0K8UTF7 A0A034VLK0 A0A0K8VA58 A0A151IPE3 A0A1I8NC53 W8C2E1 A0A154PPX2 B3NZF2 A0A3B0KCC2 A0A1B0A1P6 B4LXL4 A0A088AI67 B4PM53 A0A182J0V1 B4K9T7 B4NAX5 A0A1B0GFB1 A0A0M5JCC7 A0A1I8PII3 A0A1I8PIB7 Q293T2 A0A1A9VL97 B4GLN5 A0A164ZV96 A0A0P5CHZ9 A0A182JVH4 A0A0P5E1Z5 A0A2P8ZCQ8 A0A0P6HA89 B3LXC2 A0A1W4W1L9 E2BZA4 A0A1B0BA89 A0A0P5DVY8 A0A1A9XW62 A0A0P5RDV4 B4HI96 A0A0B4K737 Q9VGP2 A0A0P6AQK7 A0A1S3J4V6 A0A1S3J6K0 A0A1S3J508 A0A1S3J515 A0A147BDS6 A0A1S3J4W1 A0A0P5BHF8 A0A1S3J5A3 A0A0N8BZW5 A0A1S3J6K5
A0A212F8K0 A0A0L7LGH9 A0A182QPL4 A0A1Q3FSG1 A0A182GG60 A0A067RK60 A0A151WKL4 A0A182VWI7 A0A1Y9H2T1 E2AXY6 A0A232ESQ0 K7IZS1 A0A182R4V8 A0A026WKI5 A0A1S4FWX8 A0A182PGV0 A0A195ECK8 A0A182T7V6 A0A2C9GQL4 W5J6V5 A0A182UMT5 A0A0L7QPU6 Q380J0 F4WQ87 Q16LE2 A0A182L9I2 A0A1I8JVQ5 A0A158NXM3 A0A182FRF5 A0A084W244 A0A182TH75 A0A336LIB5 B0W2B2 A0A0C9QT70 A0A195EUJ9 A0A195AZZ7 A0A1B0CC23 A0A182MCE4 A0A1B0D8F4 B4JTK0 A0A0L0C7B9 A0A034VJ86 A0A0K8UTF7 A0A034VLK0 A0A0K8VA58 A0A151IPE3 A0A1I8NC53 W8C2E1 A0A154PPX2 B3NZF2 A0A3B0KCC2 A0A1B0A1P6 B4LXL4 A0A088AI67 B4PM53 A0A182J0V1 B4K9T7 B4NAX5 A0A1B0GFB1 A0A0M5JCC7 A0A1I8PII3 A0A1I8PIB7 Q293T2 A0A1A9VL97 B4GLN5 A0A164ZV96 A0A0P5CHZ9 A0A182JVH4 A0A0P5E1Z5 A0A2P8ZCQ8 A0A0P6HA89 B3LXC2 A0A1W4W1L9 E2BZA4 A0A1B0BA89 A0A0P5DVY8 A0A1A9XW62 A0A0P5RDV4 B4HI96 A0A0B4K737 Q9VGP2 A0A0P6AQK7 A0A1S3J4V6 A0A1S3J6K0 A0A1S3J508 A0A1S3J515 A0A147BDS6 A0A1S3J4W1 A0A0P5BHF8 A0A1S3J5A3 A0A0N8BZW5 A0A1S3J6K5
EC Number
1.-.-.-
Pubmed
19121390
28756777
26354079
22118469
26227816
26483478
+ More
24845553 20798317 28648823 20075255 24508170 30249741 20920257 23761445 12364791 14747013 17210077 21719571 17510324 20966253 21347285 24438588 17994087 26108605 25348373 25315136 24495485 17550304 15632085 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 15640158 16464863 29652888
24845553 20798317 28648823 20075255 24508170 30249741 20920257 23761445 12364791 14747013 17210077 21719571 17510324 20966253 21347285 24438588 17994087 26108605 25348373 25315136 24495485 17550304 15632085 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 15640158 16464863 29652888
EMBL
BABH01033122
KZ150229
PZC72014.1
NWSH01002291
PCG68660.1
RSAL01000284
+ More
RVE42984.1 ODYU01000900 SOQ36359.1 KQ460473 KPJ14483.1 AGBW02009707 OWR50072.1 JTDY01001201 KOB74557.1 AXCN02001536 GFDL01004692 JAV30353.1 JXUM01061197 JXUM01061198 JXUM01061199 KQ562139 KXJ76593.1 KK852463 KDR23413.1 KQ983001 KYQ48423.1 GL443740 EFN61689.1 NNAY01002392 OXU21379.1 KK107168 QOIP01000001 EZA56146.1 RLU26840.1 KQ979074 KYN22948.1 APCN01000881 ADMH02002131 ETN58509.1 KQ414811 KOC60579.1 AAAB01008987 EAL38574.4 GL888264 EGI63633.1 CH477910 EAT35118.1 ADTU01003167 ATLV01019496 KE525273 KFB44288.1 UFQT01000011 SSX17576.1 DS231826 EDS28727.1 GBYB01003817 JAG73584.1 KQ981958 KYN31930.1 KQ976692 KYM77781.1 AJWK01006096 AXCM01009108 AJVK01012613 CH916373 EDV95090.1 JRES01000819 KNC28170.1 GAKP01015611 JAC43341.1 GDHF01022342 GDHF01006416 JAI29972.1 JAI45898.1 GAKP01015613 JAC43339.1 GDHF01016470 JAI35844.1 KQ976877 KYN07672.1 GAMC01006589 JAB99966.1 KQ435026 KZC13943.1 CH954181 EDV49525.1 OUUW01000008 SPP83839.1 CH940650 EDW66798.1 CM000160 EDW96919.1 CH933806 EDW15585.1 CH964232 EDW80939.1 CCAG010013983 CP012526 ALC46368.1 CM000070 EAL29132.4 CH479185 EDW38459.1 LRGB01000687 KZS16816.1 GDIP01185934 JAJ37468.1 GDIP01149035 JAJ74367.1 PYGN01000102 PSN54282.1 GDIQ01022464 JAN72273.1 CH902617 EDV42766.1 GL451577 EFN79010.1 JXJN01010867 GDIP01150429 JAJ72973.1 GDIQ01102784 JAL48942.1 CH480815 EDW42611.1 AE014297 AFH06365.1 AY118818 GDIP01025849 JAM77866.1 GEGO01006493 JAR88911.1 GDIP01184679 JAJ38723.1 GDIQ01128539 JAL23187.1
RVE42984.1 ODYU01000900 SOQ36359.1 KQ460473 KPJ14483.1 AGBW02009707 OWR50072.1 JTDY01001201 KOB74557.1 AXCN02001536 GFDL01004692 JAV30353.1 JXUM01061197 JXUM01061198 JXUM01061199 KQ562139 KXJ76593.1 KK852463 KDR23413.1 KQ983001 KYQ48423.1 GL443740 EFN61689.1 NNAY01002392 OXU21379.1 KK107168 QOIP01000001 EZA56146.1 RLU26840.1 KQ979074 KYN22948.1 APCN01000881 ADMH02002131 ETN58509.1 KQ414811 KOC60579.1 AAAB01008987 EAL38574.4 GL888264 EGI63633.1 CH477910 EAT35118.1 ADTU01003167 ATLV01019496 KE525273 KFB44288.1 UFQT01000011 SSX17576.1 DS231826 EDS28727.1 GBYB01003817 JAG73584.1 KQ981958 KYN31930.1 KQ976692 KYM77781.1 AJWK01006096 AXCM01009108 AJVK01012613 CH916373 EDV95090.1 JRES01000819 KNC28170.1 GAKP01015611 JAC43341.1 GDHF01022342 GDHF01006416 JAI29972.1 JAI45898.1 GAKP01015613 JAC43339.1 GDHF01016470 JAI35844.1 KQ976877 KYN07672.1 GAMC01006589 JAB99966.1 KQ435026 KZC13943.1 CH954181 EDV49525.1 OUUW01000008 SPP83839.1 CH940650 EDW66798.1 CM000160 EDW96919.1 CH933806 EDW15585.1 CH964232 EDW80939.1 CCAG010013983 CP012526 ALC46368.1 CM000070 EAL29132.4 CH479185 EDW38459.1 LRGB01000687 KZS16816.1 GDIP01185934 JAJ37468.1 GDIP01149035 JAJ74367.1 PYGN01000102 PSN54282.1 GDIQ01022464 JAN72273.1 CH902617 EDV42766.1 GL451577 EFN79010.1 JXJN01010867 GDIP01150429 JAJ72973.1 GDIQ01102784 JAL48942.1 CH480815 EDW42611.1 AE014297 AFH06365.1 AY118818 GDIP01025849 JAM77866.1 GEGO01006493 JAR88911.1 GDIP01184679 JAJ38723.1 GDIQ01128539 JAL23187.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000007151
UP000037510
+ More
UP000075886 UP000069940 UP000249989 UP000027135 UP000075809 UP000075920 UP000075884 UP000000311 UP000215335 UP000002358 UP000075900 UP000053097 UP000279307 UP000075885 UP000078492 UP000075901 UP000075840 UP000000673 UP000075903 UP000053825 UP000007062 UP000007755 UP000008820 UP000075882 UP000076407 UP000005205 UP000069272 UP000030765 UP000075902 UP000002320 UP000078541 UP000078540 UP000092461 UP000075883 UP000092462 UP000001070 UP000037069 UP000078542 UP000095301 UP000076502 UP000008711 UP000268350 UP000092445 UP000008792 UP000005203 UP000002282 UP000075880 UP000009192 UP000007798 UP000092444 UP000092553 UP000095300 UP000001819 UP000078200 UP000008744 UP000076858 UP000075881 UP000245037 UP000007801 UP000192221 UP000008237 UP000092460 UP000092443 UP000001292 UP000000803 UP000085678
UP000075886 UP000069940 UP000249989 UP000027135 UP000075809 UP000075920 UP000075884 UP000000311 UP000215335 UP000002358 UP000075900 UP000053097 UP000279307 UP000075885 UP000078492 UP000075901 UP000075840 UP000000673 UP000075903 UP000053825 UP000007062 UP000007755 UP000008820 UP000075882 UP000076407 UP000005205 UP000069272 UP000030765 UP000075902 UP000002320 UP000078541 UP000078540 UP000092461 UP000075883 UP000092462 UP000001070 UP000037069 UP000078542 UP000095301 UP000076502 UP000008711 UP000268350 UP000092445 UP000008792 UP000005203 UP000002282 UP000075880 UP000009192 UP000007798 UP000092444 UP000092553 UP000095300 UP000001819 UP000078200 UP000008744 UP000076858 UP000075881 UP000245037 UP000007801 UP000192221 UP000008237 UP000092460 UP000092443 UP000001292 UP000000803 UP000085678
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H9JS64
A0A2W1BE30
A0A2A4JB55
A0A3S2N6W1
A0A2H1V6D6
A0A194R9G6
+ More
A0A212F8K0 A0A0L7LGH9 A0A182QPL4 A0A1Q3FSG1 A0A182GG60 A0A067RK60 A0A151WKL4 A0A182VWI7 A0A1Y9H2T1 E2AXY6 A0A232ESQ0 K7IZS1 A0A182R4V8 A0A026WKI5 A0A1S4FWX8 A0A182PGV0 A0A195ECK8 A0A182T7V6 A0A2C9GQL4 W5J6V5 A0A182UMT5 A0A0L7QPU6 Q380J0 F4WQ87 Q16LE2 A0A182L9I2 A0A1I8JVQ5 A0A158NXM3 A0A182FRF5 A0A084W244 A0A182TH75 A0A336LIB5 B0W2B2 A0A0C9QT70 A0A195EUJ9 A0A195AZZ7 A0A1B0CC23 A0A182MCE4 A0A1B0D8F4 B4JTK0 A0A0L0C7B9 A0A034VJ86 A0A0K8UTF7 A0A034VLK0 A0A0K8VA58 A0A151IPE3 A0A1I8NC53 W8C2E1 A0A154PPX2 B3NZF2 A0A3B0KCC2 A0A1B0A1P6 B4LXL4 A0A088AI67 B4PM53 A0A182J0V1 B4K9T7 B4NAX5 A0A1B0GFB1 A0A0M5JCC7 A0A1I8PII3 A0A1I8PIB7 Q293T2 A0A1A9VL97 B4GLN5 A0A164ZV96 A0A0P5CHZ9 A0A182JVH4 A0A0P5E1Z5 A0A2P8ZCQ8 A0A0P6HA89 B3LXC2 A0A1W4W1L9 E2BZA4 A0A1B0BA89 A0A0P5DVY8 A0A1A9XW62 A0A0P5RDV4 B4HI96 A0A0B4K737 Q9VGP2 A0A0P6AQK7 A0A1S3J4V6 A0A1S3J6K0 A0A1S3J508 A0A1S3J515 A0A147BDS6 A0A1S3J4W1 A0A0P5BHF8 A0A1S3J5A3 A0A0N8BZW5 A0A1S3J6K5
A0A212F8K0 A0A0L7LGH9 A0A182QPL4 A0A1Q3FSG1 A0A182GG60 A0A067RK60 A0A151WKL4 A0A182VWI7 A0A1Y9H2T1 E2AXY6 A0A232ESQ0 K7IZS1 A0A182R4V8 A0A026WKI5 A0A1S4FWX8 A0A182PGV0 A0A195ECK8 A0A182T7V6 A0A2C9GQL4 W5J6V5 A0A182UMT5 A0A0L7QPU6 Q380J0 F4WQ87 Q16LE2 A0A182L9I2 A0A1I8JVQ5 A0A158NXM3 A0A182FRF5 A0A084W244 A0A182TH75 A0A336LIB5 B0W2B2 A0A0C9QT70 A0A195EUJ9 A0A195AZZ7 A0A1B0CC23 A0A182MCE4 A0A1B0D8F4 B4JTK0 A0A0L0C7B9 A0A034VJ86 A0A0K8UTF7 A0A034VLK0 A0A0K8VA58 A0A151IPE3 A0A1I8NC53 W8C2E1 A0A154PPX2 B3NZF2 A0A3B0KCC2 A0A1B0A1P6 B4LXL4 A0A088AI67 B4PM53 A0A182J0V1 B4K9T7 B4NAX5 A0A1B0GFB1 A0A0M5JCC7 A0A1I8PII3 A0A1I8PIB7 Q293T2 A0A1A9VL97 B4GLN5 A0A164ZV96 A0A0P5CHZ9 A0A182JVH4 A0A0P5E1Z5 A0A2P8ZCQ8 A0A0P6HA89 B3LXC2 A0A1W4W1L9 E2BZA4 A0A1B0BA89 A0A0P5DVY8 A0A1A9XW62 A0A0P5RDV4 B4HI96 A0A0B4K737 Q9VGP2 A0A0P6AQK7 A0A1S3J4V6 A0A1S3J6K0 A0A1S3J508 A0A1S3J515 A0A147BDS6 A0A1S3J4W1 A0A0P5BHF8 A0A1S3J5A3 A0A0N8BZW5 A0A1S3J6K5
PDB
5OC1
E-value=9.08329e-36,
Score=378
Ontologies
GO
Topology
Subcellular location
Secreted
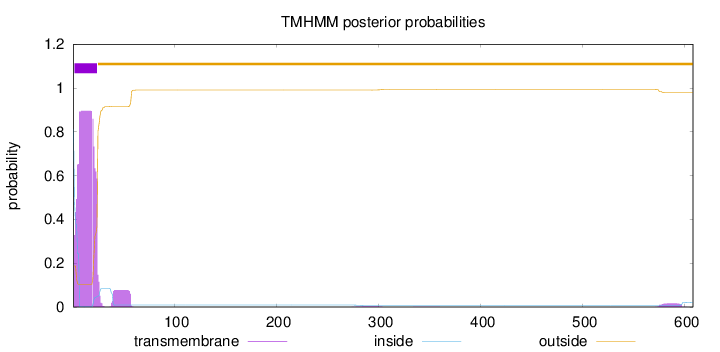
Length:
608
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.82334
Exp number, first 60 AAs:
19.39352
Total prob of N-in:
0.81232
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 24
outside
25 - 608
Population Genetic Test Statistics
Pi
186.990704
Theta
171.81243
Tajima's D
-0.59076
CLR
0.209756
CSRT
0.219389030548473
Interpretation
Uncertain
