Gene
KWMTBOMO12359
Pre Gene Modal
BGIBMGA012375
Annotation
Uncharacterized_protein_OBRU01_08898_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 0.929
Sequence
CDS
ATGTTCTCGAGTGTGAAATGCGAATCAAAAGCACGAATGGACGGCAAAACTGTCATCATAACTGGAGGCAGTGCCGGTGTTGGATTTGAAGCTGCTAAAGATTTAGCTGACAAAGGCGCTAGAGTTATCATCGCTAGTCGCAACGAAACAAAACTAAGAAAGGCTCGTGAAGAAATCATCAAGGCAACAGGAAACACTAACGTTGCGTACAAACAACTAGACCTGAGCTCTTTGAAATCAGTGCGAAATTTCGCTAACCAGATCAATACGGAAGGAAGAGTAGATGTTTTGATCAACAATGCTGGCGCGATCGCACTACCAGACGCAGTGACAACAGATAGTCTAAACCTCACCATGCAGATAAACTTTTACGGCGCTTTCTTACTCACGTTCCTATTGTTGCCAAGACTTAAAGCGTCATCTCCGAGTCGTATTATCAACGGGTCTGCGGCTGCAATGTACTTAGGAGAAATAGACTTCGATCACTGGAATGACGTTGGTCGGTATTCAATCGTGACGTCATTGGCTAACTCAAAATTAGCGATGGTCTTGTTCAATGCCGAATTGTCCAGGAGACTAGAAGGCAGTGGCGTCACTGCTAACACATACGATCCCTTCGTAGTCAAAGACACAGATATATTGACAAACCTACCTGGATTCATAAAGGAGATATCTAGATTTTTCGTGAATATAATCGGACAAGAAAAAGAAGATGCTGGTAGGCAGATATCATATTTGGCCTCCGCTCCGGAAATGGTGAATATTAGTAATCAACATTACAAGTTTTGTGAAAAATTCTTGAGGCATTGGTCGGTAAGTGATACTGGACTTACTAGAAGAGTTTGGGAAGAATCGAAGAGGCTCGTAAAAATCAGTGCAGAAGAAGACTGGGAGAGTTAG
Protein
MFSSVKCESKARMDGKTVIITGGSAGVGFEAAKDLADKGARVIIASRNETKLRKAREEIIKATGNTNVAYKQLDLSSLKSVRNFANQINTEGRVDVLINNAGAIALPDAVTTDSLNLTMQINFYGAFLLTFLLLPRLKASSPSRIINGSAAAMYLGEIDFDHWNDVGRYSIVTSLANSKLAMVLFNAELSRRLEGSGVTANTYDPFVVKDTDILTNLPGFIKEISRFFVNIIGQEKEDAGRQISYLASAPEMVNISNQHYKFCEKFLRHWSVSDTGLTRRVWEESKRLVKISAEEDWES
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9JS65
A0A2A4JUI9
A0A0L7LGP0
A0A194RAF5
A0A194PVP5
A0A2W1BFL9
+ More
A0A212F8N1 A0A2A4JWA9 A0A194RB30 A0A2W1BA63 A0A0L7LT71 A0A0N0PAN9 A0A0N0PAY1 H9JSL6 A0A2J7PGU9 A0A0L7KVJ8 A0A2H1W7C7 W4ZIF1 A0A2A4ITG8 A0A0N1IF98 A0A212F5B7 A0A0L7KV18 A0A067QUP0 A0A0N0PAB9 A0A433SWW0 A0A2J7PGT7 I4DQA1 A0A423SU57 D6X0B3 H9JSR3 A0A2J7PGT0 A0A2A4IXN8 A0A2H1W6X7 A0A087TBA3 D6X0B0 A0A1Q3G5G7 A0A2J7PGV7 B3RKN4 A0A369RY25 A0A2J7PGY1 B3SEC9 A0A0N0PAY2 C3ZKN3 B3RKN5 A0A369RLX1 B3SEC8 A0A0L7RHT2 B3RKN6 A0A369RXE5 A0A088AQ58 A0A2H1WZT2 A0A2T7P9B3 A0A1Y1N8N2 A0A0A1XDP5 A0A369RZQ8 B3RKK3 A0A369RPP6 C3XRT1 A0A131Y106 A0A0N0PB70 A0A194Q6L5 A0A1W4WGA8 A0A1Y1NCI4 A0A369RXD1 A0A131XZ97 B3RKN3 A0A0P4W6I9 A0A1W4WGK2 A0A034W5L3 A0A0K8URJ7 A0A151IL06 A0A1Z5LCY1 A0A0A1X7Y5 W8BZ25 C3YDD9 A0A069DST5 K1R790 A0A1Y1LAJ4 W4YYS4 A0A1W4WGA3 A0A151WEM9 A0A0N8E5H6 G7NN76 C3XUY5 A0A147BH14 A0A1B6DPI9 A0A0N0PC61 K1Q9G0 C3XZ20 D6X0A8 H9EVT6 A0A1U7U3F7 A0A2C9K9F2 H9H4A2 V4AKA2 E2AM66 A0A034W3X1 B4K931 A0A0K8VZD3 A0A1A9VER4
A0A212F8N1 A0A2A4JWA9 A0A194RB30 A0A2W1BA63 A0A0L7LT71 A0A0N0PAN9 A0A0N0PAY1 H9JSL6 A0A2J7PGU9 A0A0L7KVJ8 A0A2H1W7C7 W4ZIF1 A0A2A4ITG8 A0A0N1IF98 A0A212F5B7 A0A0L7KV18 A0A067QUP0 A0A0N0PAB9 A0A433SWW0 A0A2J7PGT7 I4DQA1 A0A423SU57 D6X0B3 H9JSR3 A0A2J7PGT0 A0A2A4IXN8 A0A2H1W6X7 A0A087TBA3 D6X0B0 A0A1Q3G5G7 A0A2J7PGV7 B3RKN4 A0A369RY25 A0A2J7PGY1 B3SEC9 A0A0N0PAY2 C3ZKN3 B3RKN5 A0A369RLX1 B3SEC8 A0A0L7RHT2 B3RKN6 A0A369RXE5 A0A088AQ58 A0A2H1WZT2 A0A2T7P9B3 A0A1Y1N8N2 A0A0A1XDP5 A0A369RZQ8 B3RKK3 A0A369RPP6 C3XRT1 A0A131Y106 A0A0N0PB70 A0A194Q6L5 A0A1W4WGA8 A0A1Y1NCI4 A0A369RXD1 A0A131XZ97 B3RKN3 A0A0P4W6I9 A0A1W4WGK2 A0A034W5L3 A0A0K8URJ7 A0A151IL06 A0A1Z5LCY1 A0A0A1X7Y5 W8BZ25 C3YDD9 A0A069DST5 K1R790 A0A1Y1LAJ4 W4YYS4 A0A1W4WGA3 A0A151WEM9 A0A0N8E5H6 G7NN76 C3XUY5 A0A147BH14 A0A1B6DPI9 A0A0N0PC61 K1Q9G0 C3XZ20 D6X0A8 H9EVT6 A0A1U7U3F7 A0A2C9K9F2 H9H4A2 V4AKA2 E2AM66 A0A034W3X1 B4K931 A0A0K8VZD3 A0A1A9VER4
Pubmed
EMBL
BABH01033122
NWSH01000557
PCG75671.1
JTDY01001201
KOB74559.1
KQ460473
+ More
KPJ14484.1 KQ459591 KPI97063.1 KZ150229 PZC72017.1 AGBW02009707 OWR50073.1 PCG75672.1 KPJ14485.1 PZC72018.1 JTDY01000138 KOB78650.1 KQ458665 KPJ05596.1 KQ461133 KPJ08898.1 BABH01032240 NEVH01025161 PNF15560.1 JTDY01005329 KOB67106.1 ODYU01006737 SOQ48856.1 AAGJ04078790 NWSH01008708 PCG62440.1 KPJ08897.1 AGBW02010210 OWR48927.1 KOB67107.1 KK853387 KDR07942.1 KPJ05595.1 RQTK01000904 RUS73752.1 PNF15542.1 AK404009 BAM20091.1 QCYY01002763 ROT67754.1 KQ971372 EFA10520.1 BABH01032245 PNF15543.1 NWSH01005060 PCG64471.1 SOQ48855.1 KK114425 KFM62392.1 EFA10517.1 GEYN01000196 JAV44793.1 PNF15564.1 DS985241 EDV29193.1 NOWV01000138 RDD39359.1 PNF15592.1 DS985430 EDV18917.1 KPJ08901.1 GG666638 EEN46882.1 EDV29194.1 NOWV01000972 RDD35818.1 EDV18916.1 KQ414590 KOC70369.1 EDV29195.1 NOWV01000640 RDD36014.1 RDD39358.1 ODYU01012310 SOQ58563.1 PZQS01000005 PVD30017.1 GEZM01009786 JAV94304.1 GBXI01016837 GBXI01005231 JAC97454.1 JAD09061.1 NOWV01000110 RDD40168.1 EDV29907.1 NOWV01001097 RDD35773.1 GG666456 EEN69399.1 GEFM01004225 JAP71571.1 KPJ08899.1 KQ459582 KPI99040.1 GEZM01009785 JAV94305.1 NOWV01000344 RDD36616.1 RDD39355.1 GEFM01004224 JAP71572.1 EDV29924.1 GDRN01096942 GDRN01096940 JAI59223.1 GAKP01009537 JAC49415.1 GDHF01023108 JAI29206.1 KQ977151 KYN05278.1 GFJQ02001830 JAW05140.1 GBXI01006878 JAD07414.1 GAMC01004622 JAC01934.1 GG666503 EEN61750.1 GBGD01002103 JAC86786.1 JH818186 EKC41628.1 GEZM01063043 JAV69350.1 AAGJ04004555 KQ983238 KYQ46262.1 GDIQ01059860 JAN34877.1 CM001271 EHH30398.1 GG666467 EEN68210.1 GEGO01005662 JAR89742.1 GEDC01009721 JAS27577.1 KQ460651 KPJ13037.1 JH817020 EKC30553.1 GG666475 EEN66584.1 EFA09602.1 JU322739 AFE66495.1 JSUE03022344 KB201701 ESO95165.1 GL440715 EFN65472.1 GAKP01009538 JAC49414.1 CH933806 EDW14444.1 GDHF01008354 JAI43960.1
KPJ14484.1 KQ459591 KPI97063.1 KZ150229 PZC72017.1 AGBW02009707 OWR50073.1 PCG75672.1 KPJ14485.1 PZC72018.1 JTDY01000138 KOB78650.1 KQ458665 KPJ05596.1 KQ461133 KPJ08898.1 BABH01032240 NEVH01025161 PNF15560.1 JTDY01005329 KOB67106.1 ODYU01006737 SOQ48856.1 AAGJ04078790 NWSH01008708 PCG62440.1 KPJ08897.1 AGBW02010210 OWR48927.1 KOB67107.1 KK853387 KDR07942.1 KPJ05595.1 RQTK01000904 RUS73752.1 PNF15542.1 AK404009 BAM20091.1 QCYY01002763 ROT67754.1 KQ971372 EFA10520.1 BABH01032245 PNF15543.1 NWSH01005060 PCG64471.1 SOQ48855.1 KK114425 KFM62392.1 EFA10517.1 GEYN01000196 JAV44793.1 PNF15564.1 DS985241 EDV29193.1 NOWV01000138 RDD39359.1 PNF15592.1 DS985430 EDV18917.1 KPJ08901.1 GG666638 EEN46882.1 EDV29194.1 NOWV01000972 RDD35818.1 EDV18916.1 KQ414590 KOC70369.1 EDV29195.1 NOWV01000640 RDD36014.1 RDD39358.1 ODYU01012310 SOQ58563.1 PZQS01000005 PVD30017.1 GEZM01009786 JAV94304.1 GBXI01016837 GBXI01005231 JAC97454.1 JAD09061.1 NOWV01000110 RDD40168.1 EDV29907.1 NOWV01001097 RDD35773.1 GG666456 EEN69399.1 GEFM01004225 JAP71571.1 KPJ08899.1 KQ459582 KPI99040.1 GEZM01009785 JAV94305.1 NOWV01000344 RDD36616.1 RDD39355.1 GEFM01004224 JAP71572.1 EDV29924.1 GDRN01096942 GDRN01096940 JAI59223.1 GAKP01009537 JAC49415.1 GDHF01023108 JAI29206.1 KQ977151 KYN05278.1 GFJQ02001830 JAW05140.1 GBXI01006878 JAD07414.1 GAMC01004622 JAC01934.1 GG666503 EEN61750.1 GBGD01002103 JAC86786.1 JH818186 EKC41628.1 GEZM01063043 JAV69350.1 AAGJ04004555 KQ983238 KYQ46262.1 GDIQ01059860 JAN34877.1 CM001271 EHH30398.1 GG666467 EEN68210.1 GEGO01005662 JAR89742.1 GEDC01009721 JAS27577.1 KQ460651 KPJ13037.1 JH817020 EKC30553.1 GG666475 EEN66584.1 EFA09602.1 JU322739 AFE66495.1 JSUE03022344 KB201701 ESO95165.1 GL440715 EFN65472.1 GAKP01009538 JAC49414.1 CH933806 EDW14444.1 GDHF01008354 JAI43960.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000235965 UP000007110 UP000027135 UP000271974 UP000283509 UP000007266 UP000054359 UP000009022 UP000253843 UP000001554 UP000053825 UP000005203 UP000245119 UP000192223 UP000078542 UP000005408 UP000075809 UP000189704 UP000076420 UP000006718 UP000030746 UP000000311 UP000009192 UP000078200
UP000235965 UP000007110 UP000027135 UP000271974 UP000283509 UP000007266 UP000054359 UP000009022 UP000253843 UP000001554 UP000053825 UP000005203 UP000245119 UP000192223 UP000078542 UP000005408 UP000075809 UP000189704 UP000076420 UP000006718 UP000030746 UP000000311 UP000009192 UP000078200
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JS65
A0A2A4JUI9
A0A0L7LGP0
A0A194RAF5
A0A194PVP5
A0A2W1BFL9
+ More
A0A212F8N1 A0A2A4JWA9 A0A194RB30 A0A2W1BA63 A0A0L7LT71 A0A0N0PAN9 A0A0N0PAY1 H9JSL6 A0A2J7PGU9 A0A0L7KVJ8 A0A2H1W7C7 W4ZIF1 A0A2A4ITG8 A0A0N1IF98 A0A212F5B7 A0A0L7KV18 A0A067QUP0 A0A0N0PAB9 A0A433SWW0 A0A2J7PGT7 I4DQA1 A0A423SU57 D6X0B3 H9JSR3 A0A2J7PGT0 A0A2A4IXN8 A0A2H1W6X7 A0A087TBA3 D6X0B0 A0A1Q3G5G7 A0A2J7PGV7 B3RKN4 A0A369RY25 A0A2J7PGY1 B3SEC9 A0A0N0PAY2 C3ZKN3 B3RKN5 A0A369RLX1 B3SEC8 A0A0L7RHT2 B3RKN6 A0A369RXE5 A0A088AQ58 A0A2H1WZT2 A0A2T7P9B3 A0A1Y1N8N2 A0A0A1XDP5 A0A369RZQ8 B3RKK3 A0A369RPP6 C3XRT1 A0A131Y106 A0A0N0PB70 A0A194Q6L5 A0A1W4WGA8 A0A1Y1NCI4 A0A369RXD1 A0A131XZ97 B3RKN3 A0A0P4W6I9 A0A1W4WGK2 A0A034W5L3 A0A0K8URJ7 A0A151IL06 A0A1Z5LCY1 A0A0A1X7Y5 W8BZ25 C3YDD9 A0A069DST5 K1R790 A0A1Y1LAJ4 W4YYS4 A0A1W4WGA3 A0A151WEM9 A0A0N8E5H6 G7NN76 C3XUY5 A0A147BH14 A0A1B6DPI9 A0A0N0PC61 K1Q9G0 C3XZ20 D6X0A8 H9EVT6 A0A1U7U3F7 A0A2C9K9F2 H9H4A2 V4AKA2 E2AM66 A0A034W3X1 B4K931 A0A0K8VZD3 A0A1A9VER4
A0A212F8N1 A0A2A4JWA9 A0A194RB30 A0A2W1BA63 A0A0L7LT71 A0A0N0PAN9 A0A0N0PAY1 H9JSL6 A0A2J7PGU9 A0A0L7KVJ8 A0A2H1W7C7 W4ZIF1 A0A2A4ITG8 A0A0N1IF98 A0A212F5B7 A0A0L7KV18 A0A067QUP0 A0A0N0PAB9 A0A433SWW0 A0A2J7PGT7 I4DQA1 A0A423SU57 D6X0B3 H9JSR3 A0A2J7PGT0 A0A2A4IXN8 A0A2H1W6X7 A0A087TBA3 D6X0B0 A0A1Q3G5G7 A0A2J7PGV7 B3RKN4 A0A369RY25 A0A2J7PGY1 B3SEC9 A0A0N0PAY2 C3ZKN3 B3RKN5 A0A369RLX1 B3SEC8 A0A0L7RHT2 B3RKN6 A0A369RXE5 A0A088AQ58 A0A2H1WZT2 A0A2T7P9B3 A0A1Y1N8N2 A0A0A1XDP5 A0A369RZQ8 B3RKK3 A0A369RPP6 C3XRT1 A0A131Y106 A0A0N0PB70 A0A194Q6L5 A0A1W4WGA8 A0A1Y1NCI4 A0A369RXD1 A0A131XZ97 B3RKN3 A0A0P4W6I9 A0A1W4WGK2 A0A034W5L3 A0A0K8URJ7 A0A151IL06 A0A1Z5LCY1 A0A0A1X7Y5 W8BZ25 C3YDD9 A0A069DST5 K1R790 A0A1Y1LAJ4 W4YYS4 A0A1W4WGA3 A0A151WEM9 A0A0N8E5H6 G7NN76 C3XUY5 A0A147BH14 A0A1B6DPI9 A0A0N0PC61 K1Q9G0 C3XZ20 D6X0A8 H9EVT6 A0A1U7U3F7 A0A2C9K9F2 H9H4A2 V4AKA2 E2AM66 A0A034W3X1 B4K931 A0A0K8VZD3 A0A1A9VER4
PDB
3RD5
E-value=1.42334e-18,
Score=227
Ontologies
KEGG
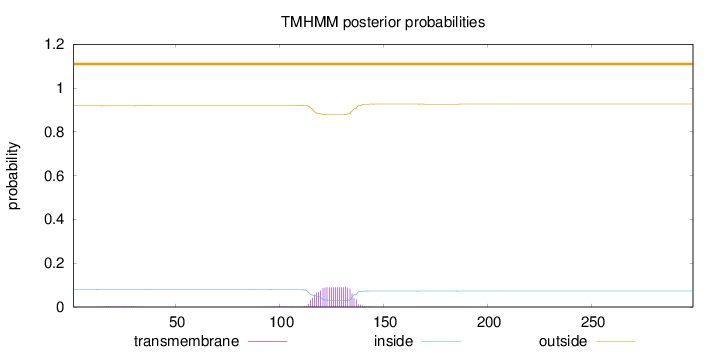
Topology
Length:
299
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.85015
Exp number, first 60 AAs:
0.02883
Total prob of N-in:
0.08095
outside
1 - 299
Population Genetic Test Statistics
Pi
79.202148
Theta
132.269096
Tajima's D
-0.979581
CLR
308.083748
CSRT
0.138993050347483
Interpretation
Uncertain
