Gene
KWMTBOMO12355
Pre Gene Modal
BGIBMGA012391
Annotation
PREDICTED:_chondroitin_sulfate_synthase_1_[Bombyx_mori]
Full name
Hexosyltransferase
+ More
Chondroitin sulfate synthase 1
Chondroitin sulfate synthase 1
Alternative Name
Chondroitin glucuronyltransferase 1
Chondroitin synthase 1
Glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase 1
N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1
N-acetylgalactosaminyltransferase 1
Chondroitin synthase 1
Glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase 1
N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1
N-acetylgalactosaminyltransferase 1
Location in the cell
Cytoplasmic Reliability : 3.141
Sequence
CDS
ATGGAGATTATTAATGCACCGTCTCGTCAACGAGGACGTGTGATTGACTTCAACGAGCTTCTGTACGGATACACCAGGCTCCAGCCGGTGCACGGGGCCGATCACATCTTGGATCTGCTGTTGAAGTACAGGAGATACAGAGGAAGGAAAATGACAGCGGCAGTTAGACGACATGCTTATTTGCAGCAGTCATTTACAGGTATGGAGATAAGAGAGTTACCAATGGGAGATCCGCCTCCATTTGAAGATCCAGCCATACTAGAAAAGACTGAGAATGGATTATATCCGAACGACTACGAGGACTTTGACGAGTCCAGTGCTCAACAGAGCAGCTTTTTGGACTTCGGTAAATTAAAAGTCAAGGAAGCTCTCGAAACTGGTCTTCAGAAGATCCAGAACAACTTACCCAAGGTGTTGAAGTGGAATAGTGATGATTCTGAAGAGTACCCGGTGTATGACAGAAAAATACACTTCGTTCTGCCCCTATCTGGTAGACAAGAGACATTCGGTAGATTCATGAAGAACTACGAAGATATAGTTTTAAAAAGCAGTCAAGCTGCCTCTCTAATTGTCGTAGCTTATTTAGATAGCAAACATCCATTGGATTATAGAAACACAGAGTTACTAATTAATTATTATATCGAGATGTACGGGAAAGACATCCAAATAGTTAAAATGGGGTCGAATATGTTTTCACGGGGAGCCGCCTTGACCGAGGGACTCAAGCTCTGCGATAGCGACGACCTTGTCTTCTTCATTGACGTGGACATGATGTTCAACCACGAGAGCCTTCGTCGAATAAGAATCAACACAATAAAATATAACCAAGTTTATTTCCCAATAGTGTTTAGTGAGTACAATCCAGATGTAGTGAACGGTGATGATTATAATAAAGTTAAAGGCGAAACTCTGTTTGTGGACACTGAATTGGAGAAGTTTATTGATGACGAAGAAACAGTGAGAACGGACAGAGAATTGGCGAATTTGAAGTACAGCCGGGAAATAACTGATGATACGGGCTATTTCAGGCAGTACGGTTTTGGTATTTTAGCTATTTATAAACGTGACTTCGAACGGATAGGGGGATTCGATTTGAACATTAAGGGTTGGGGTCTAGAAGACGTGCAGCTGTTTGAAACGCTAATTAAATCAAATTTAACGGTATACAGGATAGCTGACGAGACTCTAGTCCATATATTCCATTCGGTTGACTGTGATAAAAATTTAGAGAAGAGCCAGTTCAAGATGTGTCTGGGAACGAAGGCATCGACTTACGGCAGTGAAAAGCACATGGCGTATTATATGCTAAACCACCCTGATGTCCTGTGGCCGCCACAAGATAAACCGGAAGCGGAAGCGGAAGTGGTGGAAGAGAAACCTAAGGAAAAGAACGAGAAAGGTGCCAGGTAG
Protein
MEIINAPSRQRGRVIDFNELLYGYTRLQPVHGADHILDLLLKYRRYRGRKMTAAVRRHAYLQQSFTGMEIRELPMGDPPPFEDPAILEKTENGLYPNDYEDFDESSAQQSSFLDFGKLKVKEALETGLQKIQNNLPKVLKWNSDDSEEYPVYDRKIHFVLPLSGRQETFGRFMKNYEDIVLKSSQAASLIVVAYLDSKHPLDYRNTELLINYYIEMYGKDIQIVKMGSNMFSRGAALTEGLKLCDSDDLVFFIDVDMMFNHESLRRIRINTIKYNQVYFPIVFSEYNPDVVNGDDYNKVKGETLFVDTELEKFIDDEETVRTDRELANLKYSREITDDTGYFRQYGFGILAIYKRDFERIGGFDLNIKGWGLEDVQLFETLIKSNLTVYRIADETLVHIFHSVDCDKNLEKSQFKMCLGTKASTYGSEKHMAYYMLNHPDVLWPPQDKPEAEAEVVEEKPKEKNEKGAR
Summary
Description
Has both beta-1,3-glucuronic acid and beta-1,4-N-acetylgalactosamine transferase activity. Transfers glucuronic acid (GlcUA) from UDP-GlcUA and N-acetylgalactosamine (GalNAc) from UDP-GalNAc to the non-reducing end of the elongating chondroitin polymer.
Has both beta-1,3-glucuronic acid and beta-1,4-N-acetylgalactosamine transferase activity. Transfers glucuronic acid (GlcUA) from UDP-GlcUA and N-acetylgalactosamine (GalNAc) from UDP-GalNAc to the non-reducing end of the elongating chondroitin polymer. Involved in the negative control of osteogenesis likely through the modulation of NOTCH signaling.
Has both beta-1,3-glucuronic acid and beta-1,4-N-acetylgalactosamine transferase activity. Transfers glucuronic acid (GlcUA) from UDP-GlcUA and N-acetylgalactosamine (GalNAc) from UDP-GalNAc to the non-reducing end of the elongating chondroitin polymer. Involved in the negative control of osteogenesis likely through the modulation of NOTCH signaling.
Catalytic Activity
3-O-(beta-D-GlcA-(1->3)-beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + UDP-N-acetyl-alpha-D-galactosamine = 3-O-(beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)-beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + H(+) + UDP
3-O-(beta-D-GlcA-(1->3)-[beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)](n)-beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + UDP-N-acetyl-alpha-D-galactosamine = 3-O-([beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)](n+1)-beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + H(+) + UDP
3-O-(beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + UDP-alpha-D-glucuronate = 3-O-(beta-D-GlcA-(1->3)-beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + H(+) + UDP
3-O-([beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)](n)-beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + UDP-alpha-D-glucuronate = 3-O-(beta-D-GlcA-(1->3)-[beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)](n)-beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + H(+) + UDP
3-O-(beta-D-GlcA-(1->3)-[beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)](n)-beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + UDP-N-acetyl-alpha-D-galactosamine = 3-O-([beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)](n+1)-beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + H(+) + UDP
3-O-(beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + UDP-alpha-D-glucuronate = 3-O-(beta-D-GlcA-(1->3)-beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + H(+) + UDP
3-O-([beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)](n)-beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + UDP-alpha-D-glucuronate = 3-O-(beta-D-GlcA-(1->3)-[beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)](n)-beta-D-GalNAc-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + H(+) + UDP
Subunit
Binds CHPF.
Similarity
Belongs to the chondroitin N-acetylgalactosaminyltransferase family.
Keywords
Complete proteome
Glycoprotein
Golgi apparatus
Membrane
Metal-binding
Reference proteome
Secreted
Signal-anchor
Transferase
Transmembrane
Transmembrane helix
Disease mutation
Polymorphism
Feature
chain Hexosyltransferase
sequence variant In TPBS.
sequence variant In TPBS.
Uniprot
H9JS81
A0A1E1W9F5
A0A0L7LT67
A0A3S2NLT0
A0A2A4JUH5
A0A2W1BGU7
+ More
A0A194RA34 A0A2H1X317 A0A212F2Z6 A0A194PUH9 A0A2J7Q327 A0A2R7VP29 A0A067QLV6 A0A1B6K3F6 T1HKH9 A0A1Y1K292 A0A2P8Y5F4 A0A154PEY4 F4WTX6 A0A2J7Q328 A0A3M7PVV5 A0A0T6B2E9 A0A151JUT0 A0A151WW89 A0A3L8E3L7 A0A026VXM4 A0A158P0Y9 A0A087ZN64 A0A1S3JT82 U4UJU7 A0A0P4WQS3 A0A195AUR8 A0A151J1R6 A0A151ILV9 G3STR9 A0A3P4M7Q2 E0VDG3 A0A2Y9QQD1 V4B4G2 B0X8P8 N6U1C6 E2C9U3 A0A182H1K5 Q6ZQ11 B4DLD0 Q175Z7 A0A182KC49 A0A1S4FDT8 S9YF80 A0A0K8S8T2 H9F9J0 A0A1W4WDF8 A0A383ZW95 A0A212CSS3 L8IAE5 A0A340W9T5 A0A2K5F8I5 H2NPC0 E1ZW55 F7ALM7 A0A182MEV0 A0A1B6EH51 A0A2R9C2N3 A0A2J8VXU2 A0A2Y9GDN9 A0A2K6TUG0 A0A2K5F8K8 G3VMU5 A0A2R9C262 A0A2K6EVH1 K7C1E9 K7D3N2 A0A2I3H8Z7 Q86X52 A0A210QB45 A0A2K5RAV2 F7HY89 A0A383ZVN5 G3R0R0 W5PI31 A0A2I3GNC5 A0A1D5RD54 A0A1U7U9G8 F1SR91 A0A2K5ICG6 A0A2U4C7Z0 A0A0D9RWB0 A0A2K6R1Q3 A0A287BB21 A0A2K5YES0 A0A2Y9QGI2 K1R4W5 A0A2K6LE64 A0A2K6AQ72 A0A2K6R1L7 A0A287AWP7 A0A3Q7T9R5 G7MWB6 H0WNJ9 A0A341D052
A0A194RA34 A0A2H1X317 A0A212F2Z6 A0A194PUH9 A0A2J7Q327 A0A2R7VP29 A0A067QLV6 A0A1B6K3F6 T1HKH9 A0A1Y1K292 A0A2P8Y5F4 A0A154PEY4 F4WTX6 A0A2J7Q328 A0A3M7PVV5 A0A0T6B2E9 A0A151JUT0 A0A151WW89 A0A3L8E3L7 A0A026VXM4 A0A158P0Y9 A0A087ZN64 A0A1S3JT82 U4UJU7 A0A0P4WQS3 A0A195AUR8 A0A151J1R6 A0A151ILV9 G3STR9 A0A3P4M7Q2 E0VDG3 A0A2Y9QQD1 V4B4G2 B0X8P8 N6U1C6 E2C9U3 A0A182H1K5 Q6ZQ11 B4DLD0 Q175Z7 A0A182KC49 A0A1S4FDT8 S9YF80 A0A0K8S8T2 H9F9J0 A0A1W4WDF8 A0A383ZW95 A0A212CSS3 L8IAE5 A0A340W9T5 A0A2K5F8I5 H2NPC0 E1ZW55 F7ALM7 A0A182MEV0 A0A1B6EH51 A0A2R9C2N3 A0A2J8VXU2 A0A2Y9GDN9 A0A2K6TUG0 A0A2K5F8K8 G3VMU5 A0A2R9C262 A0A2K6EVH1 K7C1E9 K7D3N2 A0A2I3H8Z7 Q86X52 A0A210QB45 A0A2K5RAV2 F7HY89 A0A383ZVN5 G3R0R0 W5PI31 A0A2I3GNC5 A0A1D5RD54 A0A1U7U9G8 F1SR91 A0A2K5ICG6 A0A2U4C7Z0 A0A0D9RWB0 A0A2K6R1Q3 A0A287BB21 A0A2K5YES0 A0A2Y9QGI2 K1R4W5 A0A2K6LE64 A0A2K6AQ72 A0A2K6R1L7 A0A287AWP7 A0A3Q7T9R5 G7MWB6 H0WNJ9 A0A341D052
EC Number
2.4.1.-
Pubmed
19121390
26227816
28756777
26354079
22118469
24845553
+ More
28004739 29403074 21719571 30375419 30249741 24508170 21347285 23537049 20566863 23254933 20798317 26483478 14621295 17510324 23149746 26823975 25319552 22751099 17495919 22722832 21709235 11514575 10231032 12975309 15489334 12716890 12907687 21129727 25944712 21129728 28812685 25243066 22398555 20809919 17431167 25362486 22992520 30723633 22002653
28004739 29403074 21719571 30375419 30249741 24508170 21347285 23537049 20566863 23254933 20798317 26483478 14621295 17510324 23149746 26823975 25319552 22751099 17495919 22722832 21709235 11514575 10231032 12975309 15489334 12716890 12907687 21129727 25944712 21129728 28812685 25243066 22398555 20809919 17431167 25362486 22992520 30723633 22002653
EMBL
BABH01033107
BABH01033108
BABH01033109
BABH01033110
BABH01033111
BABH01033112
+ More
BABH01033113 GDQN01007563 JAT83491.1 JTDY01000138 KOB78645.1 RSAL01000284 RVE42988.1 NWSH01000557 PCG75675.1 KZ150229 PZC72020.1 KQ460473 KPJ14487.1 ODYU01013060 SOQ59721.1 AGBW02010645 OWR48109.1 KQ459591 KPI97061.1 NEVH01019070 PNF22983.1 KK854011 PTY09284.1 KK853171 KDR10328.1 GECU01001731 JAT05976.1 ACPB03002001 ACPB03002002 GEZM01096747 JAV54588.1 PYGN01000906 PSN39485.1 KQ434890 KZC10413.1 GL888345 EGI62333.1 PNF22984.1 REGN01008568 RNA03237.1 LJIG01016119 KRT81558.1 KQ981732 KYN36557.1 KQ982691 KYQ52153.1 QOIP01000001 RLU27340.1 KK107602 EZA48558.1 ADTU01005907 ADTU01005908 ADTU01005909 KB632237 ERL90461.1 GDRN01070254 JAI63913.1 KQ976738 KYM75765.1 KQ980514 KYN15733.1 KQ977086 KYN05859.1 CYRY02007166 VCW76315.1 DS235075 EEB11419.1 KB199650 ESP05353.1 DS232496 EDS42640.1 APGK01043593 APGK01043594 KB741015 ENN75350.1 GL453902 EFN75280.1 JXUM01023219 KQ560655 KXJ81415.1 AK129255 AK296943 BAG59492.1 CH477391 EAT41961.1 KB016893 EPY82795.1 GBRD01016277 GDHC01021462 GDHC01014811 JAG49549.1 JAP97166.1 JAQ03818.1 JU327543 AFE71299.1 MKHE01000013 OWK09057.1 JH881576 ELR53460.1 ABGA01296818 ABGA01296819 ABGA01296820 ABGA01296821 ABGA01296822 GL434776 EFN74564.1 AXCM01001757 GEDC01000030 JAS37268.1 AJFE02100904 AJFE02100905 AJFE02100906 NDHI03003407 PNJ62351.1 AEFK01007197 AEFK01007198 AEFK01007199 AEFK01007200 AEFK01007201 AEFK01007202 AEFK01007203 GABC01005910 GABC01005909 GABC01005908 GABC01005907 GABC01005906 GABC01005905 GABC01005904 GABF01004070 NBAG03000304 JAA05428.1 JAA18075.1 PNI43251.1 GABD01003944 GABD01003943 GABD01003942 GABD01003941 GABD01003940 GABE01010434 GABE01010433 GABE01010432 GABE01010431 GABE01010430 JAA29156.1 JAA34309.1 ADFV01131171 ADFV01131172 ADFV01131173 ADFV01131174 ADFV01131175 ADFV01131176 ADFV01131177 ADFV01131178 ADFV01131179 AB071402 AB023207 AY358529 BC046247 NEDP02004357 OWF45955.1 GAMT01009289 GAMT01009288 GAMP01007101 JAB02572.1 JAB45654.1 CABD030098189 CABD030098190 CABD030098191 AMGL01040772 AMGL01040773 AMGL01040774 JSUE03039225 JSUE03039226 AEMK02000004 AQIB01128694 AQIB01128695 JH817566 EKC36260.1 DQIR01115652 DQIR01199911 HDA71128.1 CM001259 EHH27640.1 AAQR03057961 AAQR03057962 AAQR03057963 AAQR03057964
BABH01033113 GDQN01007563 JAT83491.1 JTDY01000138 KOB78645.1 RSAL01000284 RVE42988.1 NWSH01000557 PCG75675.1 KZ150229 PZC72020.1 KQ460473 KPJ14487.1 ODYU01013060 SOQ59721.1 AGBW02010645 OWR48109.1 KQ459591 KPI97061.1 NEVH01019070 PNF22983.1 KK854011 PTY09284.1 KK853171 KDR10328.1 GECU01001731 JAT05976.1 ACPB03002001 ACPB03002002 GEZM01096747 JAV54588.1 PYGN01000906 PSN39485.1 KQ434890 KZC10413.1 GL888345 EGI62333.1 PNF22984.1 REGN01008568 RNA03237.1 LJIG01016119 KRT81558.1 KQ981732 KYN36557.1 KQ982691 KYQ52153.1 QOIP01000001 RLU27340.1 KK107602 EZA48558.1 ADTU01005907 ADTU01005908 ADTU01005909 KB632237 ERL90461.1 GDRN01070254 JAI63913.1 KQ976738 KYM75765.1 KQ980514 KYN15733.1 KQ977086 KYN05859.1 CYRY02007166 VCW76315.1 DS235075 EEB11419.1 KB199650 ESP05353.1 DS232496 EDS42640.1 APGK01043593 APGK01043594 KB741015 ENN75350.1 GL453902 EFN75280.1 JXUM01023219 KQ560655 KXJ81415.1 AK129255 AK296943 BAG59492.1 CH477391 EAT41961.1 KB016893 EPY82795.1 GBRD01016277 GDHC01021462 GDHC01014811 JAG49549.1 JAP97166.1 JAQ03818.1 JU327543 AFE71299.1 MKHE01000013 OWK09057.1 JH881576 ELR53460.1 ABGA01296818 ABGA01296819 ABGA01296820 ABGA01296821 ABGA01296822 GL434776 EFN74564.1 AXCM01001757 GEDC01000030 JAS37268.1 AJFE02100904 AJFE02100905 AJFE02100906 NDHI03003407 PNJ62351.1 AEFK01007197 AEFK01007198 AEFK01007199 AEFK01007200 AEFK01007201 AEFK01007202 AEFK01007203 GABC01005910 GABC01005909 GABC01005908 GABC01005907 GABC01005906 GABC01005905 GABC01005904 GABF01004070 NBAG03000304 JAA05428.1 JAA18075.1 PNI43251.1 GABD01003944 GABD01003943 GABD01003942 GABD01003941 GABD01003940 GABE01010434 GABE01010433 GABE01010432 GABE01010431 GABE01010430 JAA29156.1 JAA34309.1 ADFV01131171 ADFV01131172 ADFV01131173 ADFV01131174 ADFV01131175 ADFV01131176 ADFV01131177 ADFV01131178 ADFV01131179 AB071402 AB023207 AY358529 BC046247 NEDP02004357 OWF45955.1 GAMT01009289 GAMT01009288 GAMP01007101 JAB02572.1 JAB45654.1 CABD030098189 CABD030098190 CABD030098191 AMGL01040772 AMGL01040773 AMGL01040774 JSUE03039225 JSUE03039226 AEMK02000004 AQIB01128694 AQIB01128695 JH817566 EKC36260.1 DQIR01115652 DQIR01199911 HDA71128.1 CM001259 EHH27640.1 AAQR03057961 AAQR03057962 AAQR03057963 AAQR03057964
Proteomes
UP000005204
UP000037510
UP000283053
UP000218220
UP000053240
UP000007151
+ More
UP000053268 UP000235965 UP000027135 UP000015103 UP000245037 UP000076502 UP000007755 UP000276133 UP000078541 UP000075809 UP000279307 UP000053097 UP000005205 UP000005203 UP000085678 UP000030742 UP000078540 UP000078492 UP000078542 UP000007646 UP000009046 UP000248480 UP000030746 UP000002320 UP000019118 UP000008237 UP000069940 UP000249989 UP000000589 UP000008820 UP000075881 UP000192223 UP000261681 UP000265300 UP000233020 UP000001595 UP000000311 UP000002280 UP000075883 UP000240080 UP000248481 UP000233220 UP000007648 UP000233160 UP000001073 UP000005640 UP000242188 UP000233040 UP000008225 UP000001519 UP000002356 UP000006718 UP000189704 UP000008227 UP000233080 UP000245320 UP000029965 UP000233200 UP000233140 UP000248483 UP000005408 UP000233180 UP000233120 UP000286640 UP000005225 UP000252040
UP000053268 UP000235965 UP000027135 UP000015103 UP000245037 UP000076502 UP000007755 UP000276133 UP000078541 UP000075809 UP000279307 UP000053097 UP000005205 UP000005203 UP000085678 UP000030742 UP000078540 UP000078492 UP000078542 UP000007646 UP000009046 UP000248480 UP000030746 UP000002320 UP000019118 UP000008237 UP000069940 UP000249989 UP000000589 UP000008820 UP000075881 UP000192223 UP000261681 UP000265300 UP000233020 UP000001595 UP000000311 UP000002280 UP000075883 UP000240080 UP000248481 UP000233220 UP000007648 UP000233160 UP000001073 UP000005640 UP000242188 UP000233040 UP000008225 UP000001519 UP000002356 UP000006718 UP000189704 UP000008227 UP000233080 UP000245320 UP000029965 UP000233200 UP000233140 UP000248483 UP000005408 UP000233180 UP000233120 UP000286640 UP000005225 UP000252040
Pfam
Interpro
IPR029044
Nucleotide-diphossugar_trans
+ More
IPR008428 Chond_GalNAc
IPR003378 Fringe-like
IPR016186 C-type_lectin-like/link_sf
IPR001304 C-type_lectin-like
IPR018378 C-type_lectin_CS
IPR016187 CTDL_fold
IPR032675 LRR_dom_sf
IPR034182 Kexin/furin
IPR023828 Peptidase_S8_Ser-AS
IPR002884 P_dom
IPR009703 Selenoprotein_S
IPR009030 Growth_fac_rcpt_cys_sf
IPR008979 Galactose-bd-like_sf
IPR036852 Peptidase_S8/S53_dom_sf
IPR006212 Furin_repeat
IPR010909 PLAC
IPR032778 GF_recep_IV
IPR000209 Peptidase_S8/S53_dom
IPR008428 Chond_GalNAc
IPR003378 Fringe-like
IPR016186 C-type_lectin-like/link_sf
IPR001304 C-type_lectin-like
IPR018378 C-type_lectin_CS
IPR016187 CTDL_fold
IPR032675 LRR_dom_sf
IPR034182 Kexin/furin
IPR023828 Peptidase_S8_Ser-AS
IPR002884 P_dom
IPR009703 Selenoprotein_S
IPR009030 Growth_fac_rcpt_cys_sf
IPR008979 Galactose-bd-like_sf
IPR036852 Peptidase_S8/S53_dom_sf
IPR006212 Furin_repeat
IPR010909 PLAC
IPR032778 GF_recep_IV
IPR000209 Peptidase_S8/S53_dom
SUPFAM
ProteinModelPortal
H9JS81
A0A1E1W9F5
A0A0L7LT67
A0A3S2NLT0
A0A2A4JUH5
A0A2W1BGU7
+ More
A0A194RA34 A0A2H1X317 A0A212F2Z6 A0A194PUH9 A0A2J7Q327 A0A2R7VP29 A0A067QLV6 A0A1B6K3F6 T1HKH9 A0A1Y1K292 A0A2P8Y5F4 A0A154PEY4 F4WTX6 A0A2J7Q328 A0A3M7PVV5 A0A0T6B2E9 A0A151JUT0 A0A151WW89 A0A3L8E3L7 A0A026VXM4 A0A158P0Y9 A0A087ZN64 A0A1S3JT82 U4UJU7 A0A0P4WQS3 A0A195AUR8 A0A151J1R6 A0A151ILV9 G3STR9 A0A3P4M7Q2 E0VDG3 A0A2Y9QQD1 V4B4G2 B0X8P8 N6U1C6 E2C9U3 A0A182H1K5 Q6ZQ11 B4DLD0 Q175Z7 A0A182KC49 A0A1S4FDT8 S9YF80 A0A0K8S8T2 H9F9J0 A0A1W4WDF8 A0A383ZW95 A0A212CSS3 L8IAE5 A0A340W9T5 A0A2K5F8I5 H2NPC0 E1ZW55 F7ALM7 A0A182MEV0 A0A1B6EH51 A0A2R9C2N3 A0A2J8VXU2 A0A2Y9GDN9 A0A2K6TUG0 A0A2K5F8K8 G3VMU5 A0A2R9C262 A0A2K6EVH1 K7C1E9 K7D3N2 A0A2I3H8Z7 Q86X52 A0A210QB45 A0A2K5RAV2 F7HY89 A0A383ZVN5 G3R0R0 W5PI31 A0A2I3GNC5 A0A1D5RD54 A0A1U7U9G8 F1SR91 A0A2K5ICG6 A0A2U4C7Z0 A0A0D9RWB0 A0A2K6R1Q3 A0A287BB21 A0A2K5YES0 A0A2Y9QGI2 K1R4W5 A0A2K6LE64 A0A2K6AQ72 A0A2K6R1L7 A0A287AWP7 A0A3Q7T9R5 G7MWB6 H0WNJ9 A0A341D052
A0A194RA34 A0A2H1X317 A0A212F2Z6 A0A194PUH9 A0A2J7Q327 A0A2R7VP29 A0A067QLV6 A0A1B6K3F6 T1HKH9 A0A1Y1K292 A0A2P8Y5F4 A0A154PEY4 F4WTX6 A0A2J7Q328 A0A3M7PVV5 A0A0T6B2E9 A0A151JUT0 A0A151WW89 A0A3L8E3L7 A0A026VXM4 A0A158P0Y9 A0A087ZN64 A0A1S3JT82 U4UJU7 A0A0P4WQS3 A0A195AUR8 A0A151J1R6 A0A151ILV9 G3STR9 A0A3P4M7Q2 E0VDG3 A0A2Y9QQD1 V4B4G2 B0X8P8 N6U1C6 E2C9U3 A0A182H1K5 Q6ZQ11 B4DLD0 Q175Z7 A0A182KC49 A0A1S4FDT8 S9YF80 A0A0K8S8T2 H9F9J0 A0A1W4WDF8 A0A383ZW95 A0A212CSS3 L8IAE5 A0A340W9T5 A0A2K5F8I5 H2NPC0 E1ZW55 F7ALM7 A0A182MEV0 A0A1B6EH51 A0A2R9C2N3 A0A2J8VXU2 A0A2Y9GDN9 A0A2K6TUG0 A0A2K5F8K8 G3VMU5 A0A2R9C262 A0A2K6EVH1 K7C1E9 K7D3N2 A0A2I3H8Z7 Q86X52 A0A210QB45 A0A2K5RAV2 F7HY89 A0A383ZVN5 G3R0R0 W5PI31 A0A2I3GNC5 A0A1D5RD54 A0A1U7U9G8 F1SR91 A0A2K5ICG6 A0A2U4C7Z0 A0A0D9RWB0 A0A2K6R1Q3 A0A287BB21 A0A2K5YES0 A0A2Y9QGI2 K1R4W5 A0A2K6LE64 A0A2K6AQ72 A0A2K6R1L7 A0A287AWP7 A0A3Q7T9R5 G7MWB6 H0WNJ9 A0A341D052
PDB
4EEO
E-value=0.0219878,
Score=89
Ontologies
PATHWAY
GO
GO:0032580
GO:0008376
GO:0016021
GO:0016740
GO:0060349
GO:0005576
GO:0030206
GO:0051923
GO:0047238
GO:0030279
GO:0009954
GO:0002063
GO:0045880
GO:0030246
GO:0050510
GO:0016757
GO:0051216
GO:0031667
GO:0046872
GO:0006886
GO:0004252
GO:0030176
GO:0000139
GO:0016020
GO:0008270
GO:0005509
GO:0005524
GO:0003909
GO:0051103
GO:0000049
GO:0005975
GO:0004553
Topology
Subcellular location
Golgi apparatus
Golgi stack membrane
Secreted
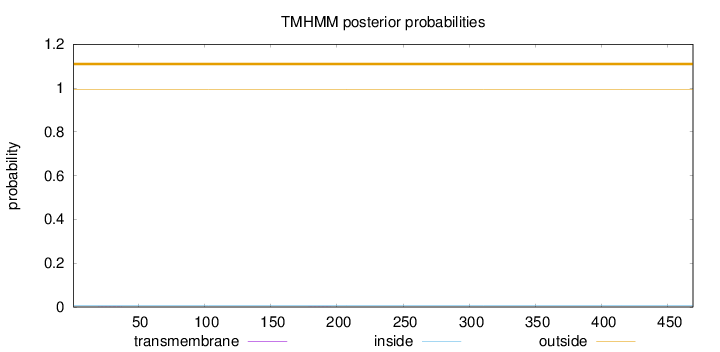
Length:
469
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00702999999999999
Exp number, first 60 AAs:
0.00354
Total prob of N-in:
0.00578
outside
1 - 469
Population Genetic Test Statistics
Pi
71.541114
Theta
141.692279
Tajima's D
-1.362199
CLR
415.174814
CSRT
0.0788460576971151
Interpretation
Uncertain
