Gene
KWMTBOMO12354
Pre Gene Modal
BGIBMGA012391
Annotation
PREDICTED:_chondroitin_sulfate_synthase_1_[Bombyx_mori]
Full name
Hexosyltransferase
Location in the cell
Cytoplasmic Reliability : 1.405 Mitochondrial Reliability : 1.99
Sequence
CDS
ATGCAAACGATTCTCCATCACAATGGCAGCGGCGAGGCTGCCTACACCGGTCGGTTGAAAGCCAGAGAGGTGCACCGAGCCATCACCTTGCATCCGGTGAAAGACCATCGCCAGATGTACAGAATACATTCATATTTCAAGAACTTTCGTATCCAAGAACTTCGCGAACGTTCCCTGGATCTCCACCGGGACATCTCCTCGGCGCTTCGGGAGCTCTCTATTCCCCGGGACGAGGTCGATGACTTTGTCTTACCCGGAGACGTCCCGCTGTTCCCGGCCAAGATTGGGGAGCCAGGATACTTGGGGGATAATAAAATATTAGGTGCCCCGATAGACATGAATCGGTACAAGCCCTCCAATCTAGACGATATCGTCATATTCGACTTCATCAGCAAGAGCCTGTACTCCGCCTACCACTCAAACCCGAAAAGACGAGTGGAGCTACCCCTGAAAGAAGCTATGGATGATGTTATCAGAGAAAAGTATAATAGTCCTTGA
Protein
MQTILHHNGSGEAAYTGRLKAREVHRAITLHPVKDHRQMYRIHSYFKNFRIQELRERSLDLHRDISSALRELSIPRDEVDDFVLPGDVPLFPAKIGEPGYLGDNKILGAPIDMNRYKPSNLDDIVIFDFISKSLYSAYHSNPKRRVELPLKEAMDDVIREKYNSP
Summary
Similarity
Belongs to the chondroitin N-acetylgalactosaminyltransferase family.
Uniprot
H9JS81
A0A2H1X317
A0A3S2NLT0
A0A1E1W9F5
A0A0L7LT67
A0A2W1BGU7
+ More
A0A194PUH9 A0A2A4JUH5 A0A194RA34 A0A212F2Z6 A0A2P8Y5F4 A0A2J7Q327 A0A182SU58 A0A067QLV6 A0A182KC49 A0A182MZU9 A0A182Q9N3 A0A182MEV0 A0A182W4Y1 A0A182XNE3 A0A182UBA5 A0A453YJK4 A0A182V1C0 A0A182Y4Y9 Q7QHY5 Q175Z7 A0A1S4FDT8 A0A182RPA9 A0A084VUT5 A0A182H1K5 A0A182J0V2 A0A182PDM9 B0X8P8 A0A2M4CNB6 A0A182FK38 A0A2M4BCY4 A0A2M4BCY8 A0A2M4BCY7 A0A2S2NW78 W5JWW3 U4UJU7 N6U1C6 J9JJR6 A0A1W4WDF8 A0A2H8TSQ0 A0A0L7R6R2 A0A2S2Q739 A0A087ZN64 A0A336LQ72 A0A151J1R6 E1ZW55 E2C9U3 A0A139WAC1 A0A224XFS5 A0A1J1HT45 A0A1J1HRF4 A0A3L8E3L7 A0A026VXM4 A0A0L0C948 A0A158P0Y9 A0A195AUR8 A0A151WW89 A0A154PEY4 A0A1B0CRF3 F4WTX6 A0A2R7VP29 A0A1S3DGU7 A0A1A9ZID4 A0A1Y1K5E5 W8CCX4 T1HKH9 A0A0C9RDW1 A0A1A9VRT9 A0A1I8PYK0 A0A1A9W8P6 A0A151JUT0 A0A1Y1K292 B4JLJ5 B4L3W6 A0A1B0D0U2 A0A0A1X398 A0A0K8UAW0 A0A3B0JF75 A0A034VQ09 Q7Z1Z0 A0A0Q9X210 B4NEC6 Q7KUZ9 B4PWP9 B4R588 A0A151ILV9 B4IK91 A0A1W4VG21 B3MZ25 A0A0Q9WN77 B4M3K3 B5DLQ0 A0A0R3NZ53 B4GWH1
A0A194PUH9 A0A2A4JUH5 A0A194RA34 A0A212F2Z6 A0A2P8Y5F4 A0A2J7Q327 A0A182SU58 A0A067QLV6 A0A182KC49 A0A182MZU9 A0A182Q9N3 A0A182MEV0 A0A182W4Y1 A0A182XNE3 A0A182UBA5 A0A453YJK4 A0A182V1C0 A0A182Y4Y9 Q7QHY5 Q175Z7 A0A1S4FDT8 A0A182RPA9 A0A084VUT5 A0A182H1K5 A0A182J0V2 A0A182PDM9 B0X8P8 A0A2M4CNB6 A0A182FK38 A0A2M4BCY4 A0A2M4BCY8 A0A2M4BCY7 A0A2S2NW78 W5JWW3 U4UJU7 N6U1C6 J9JJR6 A0A1W4WDF8 A0A2H8TSQ0 A0A0L7R6R2 A0A2S2Q739 A0A087ZN64 A0A336LQ72 A0A151J1R6 E1ZW55 E2C9U3 A0A139WAC1 A0A224XFS5 A0A1J1HT45 A0A1J1HRF4 A0A3L8E3L7 A0A026VXM4 A0A0L0C948 A0A158P0Y9 A0A195AUR8 A0A151WW89 A0A154PEY4 A0A1B0CRF3 F4WTX6 A0A2R7VP29 A0A1S3DGU7 A0A1A9ZID4 A0A1Y1K5E5 W8CCX4 T1HKH9 A0A0C9RDW1 A0A1A9VRT9 A0A1I8PYK0 A0A1A9W8P6 A0A151JUT0 A0A1Y1K292 B4JLJ5 B4L3W6 A0A1B0D0U2 A0A0A1X398 A0A0K8UAW0 A0A3B0JF75 A0A034VQ09 Q7Z1Z0 A0A0Q9X210 B4NEC6 Q7KUZ9 B4PWP9 B4R588 A0A151ILV9 B4IK91 A0A1W4VG21 B3MZ25 A0A0Q9WN77 B4M3K3 B5DLQ0 A0A0R3NZ53 B4GWH1
EC Number
2.4.1.-
Pubmed
19121390
26227816
28756777
26354079
22118469
29403074
+ More
24845553 25244985 12364791 14747013 17210077 17510324 24438588 26483478 20920257 23761445 23537049 20798317 18362917 19820115 30249741 24508170 26108605 21347285 21719571 28004739 24495485 17994087 18057021 25830018 25348373 12761549 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 23185243
24845553 25244985 12364791 14747013 17210077 17510324 24438588 26483478 20920257 23761445 23537049 20798317 18362917 19820115 30249741 24508170 26108605 21347285 21719571 28004739 24495485 17994087 18057021 25830018 25348373 12761549 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 23185243
EMBL
BABH01033107
BABH01033108
BABH01033109
BABH01033110
BABH01033111
BABH01033112
+ More
BABH01033113 ODYU01013060 SOQ59721.1 RSAL01000284 RVE42988.1 GDQN01007563 JAT83491.1 JTDY01000138 KOB78645.1 KZ150229 PZC72020.1 KQ459591 KPI97061.1 NWSH01000557 PCG75675.1 KQ460473 KPJ14487.1 AGBW02010645 OWR48109.1 PYGN01000906 PSN39485.1 NEVH01019070 PNF22983.1 KK853171 KDR10328.1 AXCN02002440 AXCM01001757 APCN01004786 AAAB01008811 EAA04965.5 CH477391 EAT41961.1 ATLV01017013 KE525142 KFB41729.1 JXUM01023219 KQ560655 KXJ81415.1 DS232496 EDS42640.1 GGFL01002654 MBW66832.1 GGFJ01001764 MBW50905.1 GGFJ01001765 MBW50906.1 GGFJ01001763 MBW50904.1 GGMR01008796 MBY21415.1 ADMH02000142 ETN67624.1 KB632237 ERL90461.1 APGK01043593 APGK01043594 KB741015 ENN75350.1 ABLF02027163 GFXV01005412 MBW17217.1 KQ414646 KOC66565.1 GGMS01004334 MBY73537.1 UFQT01000107 SSX20080.1 KQ980514 KYN15733.1 GL434776 EFN74564.1 GL453902 EFN75280.1 KQ971410 KYB24850.1 GFTR01005101 JAW11325.1 CVRI01000020 CRK90556.1 CRK90555.1 QOIP01000001 RLU27340.1 KK107602 EZA48558.1 JRES01000749 KNC28775.1 ADTU01005907 ADTU01005908 ADTU01005909 KQ976738 KYM75765.1 KQ982691 KYQ52153.1 KQ434890 KZC10413.1 AJWK01024711 AJWK01024712 AJWK01024713 GL888345 EGI62333.1 KK854011 PTY09284.1 GEZM01096748 JAV54586.1 GAMC01002945 GAMC01002943 GAMC01002941 JAC03615.1 ACPB03002001 ACPB03002002 GBYB01011267 JAG81034.1 KQ981732 KYN36557.1 GEZM01096747 JAV54588.1 CH916370 EDW00448.1 CH933810 EDW07244.2 AJVK01010068 AJVK01010069 AJVK01010070 AJVK01010071 AJVK01010072 GBXI01017355 GBXI01008976 GBXI01008799 JAC96936.1 JAD05316.1 JAD05493.1 GDHF01028545 GDHF01023372 JAI23769.1 JAI28942.1 OUUW01000003 SPP78862.1 GAKP01013566 JAC45386.1 AY241926 AAO85275.1 CH964239 KRF99571.1 EDW82095.2 AE014298 AAS65341.2 AHN59750.1 CM000162 EDX01795.1 CM000366 EDX17989.1 KQ977086 KYN05859.1 CH480852 EDW51483.1 CH902632 EDV32869.2 CH940651 KRF82203.1 EDW65378.1 CH379063 EDY72452.2 KRT06305.1 CH479194 EDW27055.1
BABH01033113 ODYU01013060 SOQ59721.1 RSAL01000284 RVE42988.1 GDQN01007563 JAT83491.1 JTDY01000138 KOB78645.1 KZ150229 PZC72020.1 KQ459591 KPI97061.1 NWSH01000557 PCG75675.1 KQ460473 KPJ14487.1 AGBW02010645 OWR48109.1 PYGN01000906 PSN39485.1 NEVH01019070 PNF22983.1 KK853171 KDR10328.1 AXCN02002440 AXCM01001757 APCN01004786 AAAB01008811 EAA04965.5 CH477391 EAT41961.1 ATLV01017013 KE525142 KFB41729.1 JXUM01023219 KQ560655 KXJ81415.1 DS232496 EDS42640.1 GGFL01002654 MBW66832.1 GGFJ01001764 MBW50905.1 GGFJ01001765 MBW50906.1 GGFJ01001763 MBW50904.1 GGMR01008796 MBY21415.1 ADMH02000142 ETN67624.1 KB632237 ERL90461.1 APGK01043593 APGK01043594 KB741015 ENN75350.1 ABLF02027163 GFXV01005412 MBW17217.1 KQ414646 KOC66565.1 GGMS01004334 MBY73537.1 UFQT01000107 SSX20080.1 KQ980514 KYN15733.1 GL434776 EFN74564.1 GL453902 EFN75280.1 KQ971410 KYB24850.1 GFTR01005101 JAW11325.1 CVRI01000020 CRK90556.1 CRK90555.1 QOIP01000001 RLU27340.1 KK107602 EZA48558.1 JRES01000749 KNC28775.1 ADTU01005907 ADTU01005908 ADTU01005909 KQ976738 KYM75765.1 KQ982691 KYQ52153.1 KQ434890 KZC10413.1 AJWK01024711 AJWK01024712 AJWK01024713 GL888345 EGI62333.1 KK854011 PTY09284.1 GEZM01096748 JAV54586.1 GAMC01002945 GAMC01002943 GAMC01002941 JAC03615.1 ACPB03002001 ACPB03002002 GBYB01011267 JAG81034.1 KQ981732 KYN36557.1 GEZM01096747 JAV54588.1 CH916370 EDW00448.1 CH933810 EDW07244.2 AJVK01010068 AJVK01010069 AJVK01010070 AJVK01010071 AJVK01010072 GBXI01017355 GBXI01008976 GBXI01008799 JAC96936.1 JAD05316.1 JAD05493.1 GDHF01028545 GDHF01023372 JAI23769.1 JAI28942.1 OUUW01000003 SPP78862.1 GAKP01013566 JAC45386.1 AY241926 AAO85275.1 CH964239 KRF99571.1 EDW82095.2 AE014298 AAS65341.2 AHN59750.1 CM000162 EDX01795.1 CM000366 EDX17989.1 KQ977086 KYN05859.1 CH480852 EDW51483.1 CH902632 EDV32869.2 CH940651 KRF82203.1 EDW65378.1 CH379063 EDY72452.2 KRT06305.1 CH479194 EDW27055.1
Proteomes
UP000005204
UP000283053
UP000037510
UP000053268
UP000218220
UP000053240
+ More
UP000007151 UP000245037 UP000235965 UP000075901 UP000027135 UP000075881 UP000075884 UP000075886 UP000075883 UP000075920 UP000076407 UP000075902 UP000075840 UP000075903 UP000076408 UP000007062 UP000008820 UP000075900 UP000030765 UP000069940 UP000249989 UP000075880 UP000075885 UP000002320 UP000069272 UP000000673 UP000030742 UP000019118 UP000007819 UP000192223 UP000053825 UP000005203 UP000078492 UP000000311 UP000008237 UP000007266 UP000183832 UP000279307 UP000053097 UP000037069 UP000005205 UP000078540 UP000075809 UP000076502 UP000092461 UP000007755 UP000079169 UP000092445 UP000015103 UP000078200 UP000095300 UP000091820 UP000078541 UP000001070 UP000009192 UP000092462 UP000268350 UP000007798 UP000000803 UP000002282 UP000000304 UP000078542 UP000001292 UP000192221 UP000007801 UP000008792 UP000001819 UP000008744
UP000007151 UP000245037 UP000235965 UP000075901 UP000027135 UP000075881 UP000075884 UP000075886 UP000075883 UP000075920 UP000076407 UP000075902 UP000075840 UP000075903 UP000076408 UP000007062 UP000008820 UP000075900 UP000030765 UP000069940 UP000249989 UP000075880 UP000075885 UP000002320 UP000069272 UP000000673 UP000030742 UP000019118 UP000007819 UP000192223 UP000053825 UP000005203 UP000078492 UP000000311 UP000008237 UP000007266 UP000183832 UP000279307 UP000053097 UP000037069 UP000005205 UP000078540 UP000075809 UP000076502 UP000092461 UP000007755 UP000079169 UP000092445 UP000015103 UP000078200 UP000095300 UP000091820 UP000078541 UP000001070 UP000009192 UP000092462 UP000268350 UP000007798 UP000000803 UP000002282 UP000000304 UP000078542 UP000001292 UP000192221 UP000007801 UP000008792 UP000001819 UP000008744
Pfam
Interpro
IPR029044
Nucleotide-diphossugar_trans
+ More
IPR008428 Chond_GalNAc
IPR016186 C-type_lectin-like/link_sf
IPR001304 C-type_lectin-like
IPR018378 C-type_lectin_CS
IPR016187 CTDL_fold
IPR002014 VHS_dom
IPR004152 GAT_dom
IPR008153 GAE_dom
IPR008942 ENTH_VHS
IPR008152 Clathrin_a/b/g-adaptin_app_Ig
IPR038425 GAT_sf
IPR013041 Clathrin_app_Ig-like_sf
IPR041198 GGA_N-GAT
IPR008428 Chond_GalNAc
IPR016186 C-type_lectin-like/link_sf
IPR001304 C-type_lectin-like
IPR018378 C-type_lectin_CS
IPR016187 CTDL_fold
IPR002014 VHS_dom
IPR004152 GAT_dom
IPR008153 GAE_dom
IPR008942 ENTH_VHS
IPR008152 Clathrin_a/b/g-adaptin_app_Ig
IPR038425 GAT_sf
IPR013041 Clathrin_app_Ig-like_sf
IPR041198 GGA_N-GAT
Gene 3D
ProteinModelPortal
H9JS81
A0A2H1X317
A0A3S2NLT0
A0A1E1W9F5
A0A0L7LT67
A0A2W1BGU7
+ More
A0A194PUH9 A0A2A4JUH5 A0A194RA34 A0A212F2Z6 A0A2P8Y5F4 A0A2J7Q327 A0A182SU58 A0A067QLV6 A0A182KC49 A0A182MZU9 A0A182Q9N3 A0A182MEV0 A0A182W4Y1 A0A182XNE3 A0A182UBA5 A0A453YJK4 A0A182V1C0 A0A182Y4Y9 Q7QHY5 Q175Z7 A0A1S4FDT8 A0A182RPA9 A0A084VUT5 A0A182H1K5 A0A182J0V2 A0A182PDM9 B0X8P8 A0A2M4CNB6 A0A182FK38 A0A2M4BCY4 A0A2M4BCY8 A0A2M4BCY7 A0A2S2NW78 W5JWW3 U4UJU7 N6U1C6 J9JJR6 A0A1W4WDF8 A0A2H8TSQ0 A0A0L7R6R2 A0A2S2Q739 A0A087ZN64 A0A336LQ72 A0A151J1R6 E1ZW55 E2C9U3 A0A139WAC1 A0A224XFS5 A0A1J1HT45 A0A1J1HRF4 A0A3L8E3L7 A0A026VXM4 A0A0L0C948 A0A158P0Y9 A0A195AUR8 A0A151WW89 A0A154PEY4 A0A1B0CRF3 F4WTX6 A0A2R7VP29 A0A1S3DGU7 A0A1A9ZID4 A0A1Y1K5E5 W8CCX4 T1HKH9 A0A0C9RDW1 A0A1A9VRT9 A0A1I8PYK0 A0A1A9W8P6 A0A151JUT0 A0A1Y1K292 B4JLJ5 B4L3W6 A0A1B0D0U2 A0A0A1X398 A0A0K8UAW0 A0A3B0JF75 A0A034VQ09 Q7Z1Z0 A0A0Q9X210 B4NEC6 Q7KUZ9 B4PWP9 B4R588 A0A151ILV9 B4IK91 A0A1W4VG21 B3MZ25 A0A0Q9WN77 B4M3K3 B5DLQ0 A0A0R3NZ53 B4GWH1
A0A194PUH9 A0A2A4JUH5 A0A194RA34 A0A212F2Z6 A0A2P8Y5F4 A0A2J7Q327 A0A182SU58 A0A067QLV6 A0A182KC49 A0A182MZU9 A0A182Q9N3 A0A182MEV0 A0A182W4Y1 A0A182XNE3 A0A182UBA5 A0A453YJK4 A0A182V1C0 A0A182Y4Y9 Q7QHY5 Q175Z7 A0A1S4FDT8 A0A182RPA9 A0A084VUT5 A0A182H1K5 A0A182J0V2 A0A182PDM9 B0X8P8 A0A2M4CNB6 A0A182FK38 A0A2M4BCY4 A0A2M4BCY8 A0A2M4BCY7 A0A2S2NW78 W5JWW3 U4UJU7 N6U1C6 J9JJR6 A0A1W4WDF8 A0A2H8TSQ0 A0A0L7R6R2 A0A2S2Q739 A0A087ZN64 A0A336LQ72 A0A151J1R6 E1ZW55 E2C9U3 A0A139WAC1 A0A224XFS5 A0A1J1HT45 A0A1J1HRF4 A0A3L8E3L7 A0A026VXM4 A0A0L0C948 A0A158P0Y9 A0A195AUR8 A0A151WW89 A0A154PEY4 A0A1B0CRF3 F4WTX6 A0A2R7VP29 A0A1S3DGU7 A0A1A9ZID4 A0A1Y1K5E5 W8CCX4 T1HKH9 A0A0C9RDW1 A0A1A9VRT9 A0A1I8PYK0 A0A1A9W8P6 A0A151JUT0 A0A1Y1K292 B4JLJ5 B4L3W6 A0A1B0D0U2 A0A0A1X398 A0A0K8UAW0 A0A3B0JF75 A0A034VQ09 Q7Z1Z0 A0A0Q9X210 B4NEC6 Q7KUZ9 B4PWP9 B4R588 A0A151ILV9 B4IK91 A0A1W4VG21 B3MZ25 A0A0Q9WN77 B4M3K3 B5DLQ0 A0A0R3NZ53 B4GWH1
Ontologies
GO
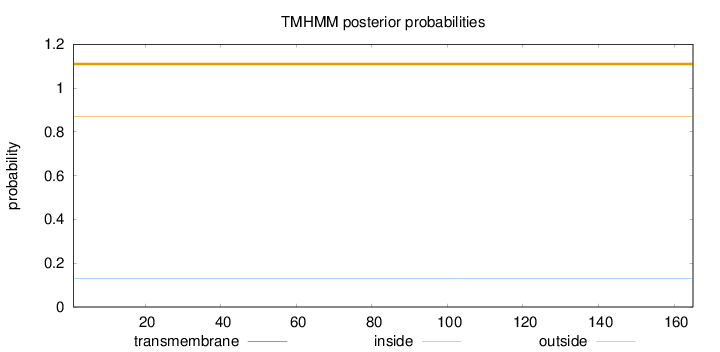
Topology
Subcellular location
Length:
165
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00146
Exp number, first 60 AAs:
0
Total prob of N-in:
0.12900
outside
1 - 165
Population Genetic Test Statistics
Pi
248.662553
Theta
19.223028
Tajima's D
0.179582
CLR
0.440776
CSRT
0.418479076046198
Interpretation
Uncertain
