Gene
KWMTBOMO12351
Pre Gene Modal
BGIBMGA012377
Annotation
PREDICTED:_poly(A)_RNA_polymerase_gld-2_homolog_A-like_isoform_X2_[Bombyx_mori]
Full name
Poly(A) RNA polymerase gld-2 homolog A
Location in the cell
Nuclear Reliability : 2.893
Sequence
CDS
ATGAACGAGGGTGCGCCTGTAATGTTCCATGCAGGCGGTGGGTACGGTCCGGGTGTTTACGTGATGCCGATGGAACGGCCTCGGCAGTATGGGCTGGAGGTGGTTCCTACGAACGTGACTGGCACAGCTTCCCCCTCGACCCCGAGTACTCCACGTCGCTGGCAGCGCCGCGACCGACAGAAGGATACAGCATTTGGATATGATTCAGATGACTCCAGTTTGTCTAGCAATGCTTCTAGCTCCAACAAGTCTGGTGAAAAAGGGGAATGTACGTCTAGCAACCCAGATCACACGGCAGCGGGGAACACGGCCCAGGTTCTGTACAACGCGCGGGAGTGGCGCGGCCGGCGGCGGGACGTGAGGCCGCAGCGCCGACTCGCACCGGAGCGGTTCTTTAAGGCCACCTATCCCTTTCAAGTTAAGTTCACACCGGATGACCTTTTAAATGGGTCCAAGTACGACACCCTGTCGCAGGAGATCTGGGACAAGTTCGTCAAGTCGCAGCAGACTGAGGAGACGTTCCGGAAGAAGATGAACCTTTGGAGATATCTATATACGATATTCCCCCGCTACGGGCTGTACGTGGTGGGCTCCACCATGTCGGGCTTCGGGCTGGAGTCGTCGGACATGGACCTGTGCCTGCACGTGCGCGCGCACCAGCACGTGGAGCCGCGCCCGCACGCGCTGCTGCACCTCAACTACATCCTCGGCTACATCCGGAGCTTCGACCCCGGTGCGGAGCTGATCCAGGCCAAGGTTCCGATCCTGAAGTTCCGCGACGTCCGCAACGGGCTCCAGGTGGACCTCAACTGCAACAACGTGGTCGGCATCAGGAACACCGACCTGCTGCACTGCTTCTCCAGAATGGACTGGCGCGTGCGGCCGCTGGTGGCGCTGGCCAAGCTGTGGGCGCGCGCGCACGCCATCAACGACGCGCGCCAGCGGACGCTCTCCTCCTACGCGCTCACGCTCATGGTCATCCACTTCCTGCAGTGCGGCACGAGCCCCCCCATCCTGCCGCGCGCCTGCGACGTGCTGCGCGCGCCCCCCCGCGGGCCCCCCGCCGCGGCCCCCCCGCACAGCCGCGCCACGCTGGGGGAGCTGTTCCTCAACATGCTGCGCTACTACGCCACCTTCCCGTACTCGCGCATGGCGGTGTCGGTCCGGGCGGGCGCGCGCGTGTGCGTGTCGGAGTGCCGCGCCGCCCGCGCCTGCAAGAACGACCCGCACCAGTGGAAGCTGCTCTGCGTCGAAGAGCCCTTCGACCTGACCAACGCGGCGCGCTCCGTGTACGACCCGGACACGTTCGAGCGCATCGTGAGCACGTTCCGGGACAGCTACAGGCGCCTGTCCGCCGGCCTGCGCCTCGGCGACGCCTGGCCGCAGCGATGA
Protein
MNEGAPVMFHAGGGYGPGVYVMPMERPRQYGLEVVPTNVTGTASPSTPSTPRRWQRRDRQKDTAFGYDSDDSSLSSNASSSNKSGEKGECTSSNPDHTAAGNTAQVLYNAREWRGRRRDVRPQRRLAPERFFKATYPFQVKFTPDDLLNGSKYDTLSQEIWDKFVKSQQTEETFRKKMNLWRYLYTIFPRYGLYVVGSTMSGFGLESSDMDLCLHVRAHQHVEPRPHALLHLNYILGYIRSFDPGAELIQAKVPILKFRDVRNGLQVDLNCNNVVGIRNTDLLHCFSRMDWRVRPLVALAKLWARAHAINDARQRTLSSYALTLMVIHFLQCGTSPPILPRACDVLRAPPRGPPAAAPPHSRATLGELFLNMLRYYATFPYSRMAVSVRAGARVCVSECRAARACKNDPHQWKLLCVEEPFDLTNAARSVYDPDTFERIVSTFRDSYRRLSAGLRLGDAWPQR
Summary
Description
Cytoplasmic poly(A) RNA polymerase that adds successive AMP monomers to the 3'-end of specific RNAs, forming a poly(A) tail. In contrast to the canonical nuclear poly(A) RNA polymerase, it only adds poly(A) to selected cytoplasmic mRNAs. Required for formation of long term memory.
Catalytic Activity
ATP + RNA(n) = diphosphate + RNA(n)-3'-adenine ribonucleotide
Subunit
Interacts with Fmr1 and eIF-4E.
Similarity
Belongs to the DNA polymerase type-B-like family. GLD2 subfamily.
Keywords
Alternative splicing
ATP-binding
Complete proteome
Cytoplasm
Magnesium
Manganese
Metal-binding
mRNA processing
Nucleotide-binding
Nucleus
Reference proteome
RNA-binding
Transferase
Feature
chain Poly(A) RNA polymerase gld-2 homolog A
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A0A2W1B8M0
A0A2H1W1W1
A0A212FBD5
A0A194PUQ5
A0A194RAG0
A0A2A4JUG2
+ More
A0A437AXT4 A0A088A209 A0A0L7QUF1 A0A154PRA7 A0A0N0BJF6 A0A067QPB4 A0A0N7Z8C7 A0A336KST5 A0A0V0G8I2 A0A023EZJ1 A0A034V9L3 A0A3L8DZ32 A0A224XA48 A0A2J7RCK1 A0A026W464 A0A1B6CDZ4 K7INU7 E0VRI1 A0A0L0BLB6 A0A182NR07 A0A1I8PZD2 A0A182YNB1 A0A0A1X667 A0A1I8MZQ6 A0A182QVW4 A0A240SWJ3 A0A0K8U9N1 A0A182RG15 A0A182VW86 Q7PWZ2 A0A182M491 E9J500 A0A3F2YZN1 A0A182JXI5 A0A182XNY6 A0A182L8B3 A0A1S4GBP6 A0A182HK32 A0A182VC05 A0A1A9W310 W5J5B6 A0A2C9GGW2 A0A151X421 A0A182PR64 A0A1D2N5Q8 A0A195EYS6 A0A195BPC8 A0A158NDR4 A0A195D7M0 F4W842 A0A1A9VU92 A0A1B0B5Y4 A0A0T6ATH7 A0A1B0EYY0 A0A1B0GL72 A0A1B0FM65 D6WCT7 A0A2P8YDJ4 A0A182IRZ8 A0A1Q3EYT2 E2A0N9 A0A1A9ZK21 A0A0A9ZCX4 B4K6N0 A0A0A9Z328 A0A1B6KX76 T1HJ60 A0A1Y1K7Q4 A0A0K8TH28 N6UST1 A0A0Q9WR14 Q176V1 A0A1S4FD04 A0A182GEY8 B4QZX8 B4HE49 A0A1B6FVA1 B3P7M5 A0A3B0JG78 B4JRM7 B4G5B6 I5ANR3 B4PLL3 B4NHS4 A0A0P5L4A4 A0A0B4KGP2 Q9VD44 A0A1W4VPD4
A0A437AXT4 A0A088A209 A0A0L7QUF1 A0A154PRA7 A0A0N0BJF6 A0A067QPB4 A0A0N7Z8C7 A0A336KST5 A0A0V0G8I2 A0A023EZJ1 A0A034V9L3 A0A3L8DZ32 A0A224XA48 A0A2J7RCK1 A0A026W464 A0A1B6CDZ4 K7INU7 E0VRI1 A0A0L0BLB6 A0A182NR07 A0A1I8PZD2 A0A182YNB1 A0A0A1X667 A0A1I8MZQ6 A0A182QVW4 A0A240SWJ3 A0A0K8U9N1 A0A182RG15 A0A182VW86 Q7PWZ2 A0A182M491 E9J500 A0A3F2YZN1 A0A182JXI5 A0A182XNY6 A0A182L8B3 A0A1S4GBP6 A0A182HK32 A0A182VC05 A0A1A9W310 W5J5B6 A0A2C9GGW2 A0A151X421 A0A182PR64 A0A1D2N5Q8 A0A195EYS6 A0A195BPC8 A0A158NDR4 A0A195D7M0 F4W842 A0A1A9VU92 A0A1B0B5Y4 A0A0T6ATH7 A0A1B0EYY0 A0A1B0GL72 A0A1B0FM65 D6WCT7 A0A2P8YDJ4 A0A182IRZ8 A0A1Q3EYT2 E2A0N9 A0A1A9ZK21 A0A0A9ZCX4 B4K6N0 A0A0A9Z328 A0A1B6KX76 T1HJ60 A0A1Y1K7Q4 A0A0K8TH28 N6UST1 A0A0Q9WR14 Q176V1 A0A1S4FD04 A0A182GEY8 B4QZX8 B4HE49 A0A1B6FVA1 B3P7M5 A0A3B0JG78 B4JRM7 B4G5B6 I5ANR3 B4PLL3 B4NHS4 A0A0P5L4A4 A0A0B4KGP2 Q9VD44 A0A1W4VPD4
EC Number
2.7.7.19
Pubmed
28756777
22118469
26354079
24845553
27129103
25474469
+ More
25348373 30249741 24508170 20075255 20566863 26108605 25244985 25830018 25315136 12364791 21282665 20966253 20920257 23761445 27289101 21347285 21719571 18362917 19820115 29403074 20798317 25401762 17994087 28004739 23537049 17510324 26483478 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 18780789
25348373 30249741 24508170 20075255 20566863 26108605 25244985 25830018 25315136 12364791 21282665 20966253 20920257 23761445 27289101 21347285 21719571 18362917 19820115 29403074 20798317 25401762 17994087 28004739 23537049 17510324 26483478 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 18780789
EMBL
KZ150229
PZC72022.1
ODYU01005550
SOQ46504.1
AGBW02009347
OWR51040.1
+ More
KQ459591 KPI97057.1 KQ460473 KPJ14489.1 NWSH01000557 PCG75677.1 RSAL01000284 RVE42990.1 KQ414735 KOC62287.1 KQ435078 KZC14446.1 KQ435716 KOX79068.1 KK853171 KDR10315.1 GDKW01003715 JAI52880.1 UFQS01000733 UFQT01000733 SSX06465.1 SSX26814.1 GECL01002429 JAP03695.1 GBBI01004045 JAC14667.1 GAKP01018946 JAC40006.1 QOIP01000002 RLU25606.1 GFTR01007200 JAW09226.1 NEVH01005885 PNF38552.1 KK107436 EZA50842.1 GEDC01025677 JAS11621.1 DS235478 EEB15987.1 JRES01001701 KNC20837.1 GBXI01016255 GBXI01008084 GBXI01007655 JAC98036.1 JAD06208.1 JAD06637.1 AXCN02001485 GDHF01029249 GDHF01002494 JAI23065.1 JAI49820.1 AAAB01008987 EAA01442.4 AXCM01000086 GL768123 EFZ12054.1 APCN01000816 ADMH02002101 ETN59176.1 KQ982548 KYQ55175.1 LJIJ01000197 ODN00609.1 KQ981906 KYN33281.1 KQ976424 KYM88372.1 ADTU01012805 KQ981153 KYN08871.1 GL887888 EGI69712.1 JXJN01008910 LJIG01022895 KRT78181.1 AJVK01006447 AJVK01006448 AJWK01034447 AJWK01034448 AJWK01034449 AJWK01034450 CCAG010007701 KQ971311 EEZ98847.1 PYGN01000679 PSN42330.1 GFDL01014596 JAV20449.1 GL435626 EFN72942.1 GBHO01003969 JAG39635.1 CH933806 EDW16330.2 GBHO01003967 JAG39637.1 GEBQ01023945 GEBQ01012682 JAT16032.1 JAT27295.1 ACPB03021239 GEZM01094313 JAV55715.1 GBRD01001130 JAG64691.1 APGK01017822 KB740053 ENN81807.1 CH940650 KRF83236.1 CH477382 EAT42185.1 JXUM01058883 JXUM01058884 JXUM01058885 KQ562023 KXJ76885.1 CM000364 EDX13884.1 CH480815 EDW43146.1 GECZ01015718 JAS54051.1 CH954182 EDV54186.1 OUUW01000005 SPP81347.1 CH916373 EDV94417.1 CH479179 EDW24782.1 CM000070 EIM52598.2 CM000160 EDW97962.1 CH964272 EDW84684.2 GDIQ01180614 JAK71111.1 AE014297 AGB96209.1 AY122076 BT044483
KQ459591 KPI97057.1 KQ460473 KPJ14489.1 NWSH01000557 PCG75677.1 RSAL01000284 RVE42990.1 KQ414735 KOC62287.1 KQ435078 KZC14446.1 KQ435716 KOX79068.1 KK853171 KDR10315.1 GDKW01003715 JAI52880.1 UFQS01000733 UFQT01000733 SSX06465.1 SSX26814.1 GECL01002429 JAP03695.1 GBBI01004045 JAC14667.1 GAKP01018946 JAC40006.1 QOIP01000002 RLU25606.1 GFTR01007200 JAW09226.1 NEVH01005885 PNF38552.1 KK107436 EZA50842.1 GEDC01025677 JAS11621.1 DS235478 EEB15987.1 JRES01001701 KNC20837.1 GBXI01016255 GBXI01008084 GBXI01007655 JAC98036.1 JAD06208.1 JAD06637.1 AXCN02001485 GDHF01029249 GDHF01002494 JAI23065.1 JAI49820.1 AAAB01008987 EAA01442.4 AXCM01000086 GL768123 EFZ12054.1 APCN01000816 ADMH02002101 ETN59176.1 KQ982548 KYQ55175.1 LJIJ01000197 ODN00609.1 KQ981906 KYN33281.1 KQ976424 KYM88372.1 ADTU01012805 KQ981153 KYN08871.1 GL887888 EGI69712.1 JXJN01008910 LJIG01022895 KRT78181.1 AJVK01006447 AJVK01006448 AJWK01034447 AJWK01034448 AJWK01034449 AJWK01034450 CCAG010007701 KQ971311 EEZ98847.1 PYGN01000679 PSN42330.1 GFDL01014596 JAV20449.1 GL435626 EFN72942.1 GBHO01003969 JAG39635.1 CH933806 EDW16330.2 GBHO01003967 JAG39637.1 GEBQ01023945 GEBQ01012682 JAT16032.1 JAT27295.1 ACPB03021239 GEZM01094313 JAV55715.1 GBRD01001130 JAG64691.1 APGK01017822 KB740053 ENN81807.1 CH940650 KRF83236.1 CH477382 EAT42185.1 JXUM01058883 JXUM01058884 JXUM01058885 KQ562023 KXJ76885.1 CM000364 EDX13884.1 CH480815 EDW43146.1 GECZ01015718 JAS54051.1 CH954182 EDV54186.1 OUUW01000005 SPP81347.1 CH916373 EDV94417.1 CH479179 EDW24782.1 CM000070 EIM52598.2 CM000160 EDW97962.1 CH964272 EDW84684.2 GDIQ01180614 JAK71111.1 AE014297 AGB96209.1 AY122076 BT044483
Proteomes
UP000007151
UP000053268
UP000053240
UP000218220
UP000283053
UP000005203
+ More
UP000053825 UP000076502 UP000053105 UP000027135 UP000279307 UP000235965 UP000053097 UP000002358 UP000009046 UP000037069 UP000075884 UP000095300 UP000076408 UP000095301 UP000075886 UP000092443 UP000075900 UP000075920 UP000007062 UP000075883 UP000075902 UP000075881 UP000076407 UP000075882 UP000075840 UP000075903 UP000091820 UP000000673 UP000069272 UP000075809 UP000075885 UP000094527 UP000078541 UP000078540 UP000005205 UP000078492 UP000007755 UP000078200 UP000092460 UP000092462 UP000092461 UP000092444 UP000007266 UP000245037 UP000075880 UP000000311 UP000092445 UP000009192 UP000015103 UP000019118 UP000008792 UP000008820 UP000069940 UP000249989 UP000000304 UP000001292 UP000008711 UP000268350 UP000001070 UP000008744 UP000001819 UP000002282 UP000007798 UP000000803 UP000192221
UP000053825 UP000076502 UP000053105 UP000027135 UP000279307 UP000235965 UP000053097 UP000002358 UP000009046 UP000037069 UP000075884 UP000095300 UP000076408 UP000095301 UP000075886 UP000092443 UP000075900 UP000075920 UP000007062 UP000075883 UP000075902 UP000075881 UP000076407 UP000075882 UP000075840 UP000075903 UP000091820 UP000000673 UP000069272 UP000075809 UP000075885 UP000094527 UP000078541 UP000078540 UP000005205 UP000078492 UP000007755 UP000078200 UP000092460 UP000092462 UP000092461 UP000092444 UP000007266 UP000245037 UP000075880 UP000000311 UP000092445 UP000009192 UP000015103 UP000019118 UP000008792 UP000008820 UP000069940 UP000249989 UP000000304 UP000001292 UP000008711 UP000268350 UP000001070 UP000008744 UP000001819 UP000002282 UP000007798 UP000000803 UP000192221
PRIDE
Interpro
SUPFAM
SSF47769
SSF47769
Gene 3D
ProteinModelPortal
A0A2W1B8M0
A0A2H1W1W1
A0A212FBD5
A0A194PUQ5
A0A194RAG0
A0A2A4JUG2
+ More
A0A437AXT4 A0A088A209 A0A0L7QUF1 A0A154PRA7 A0A0N0BJF6 A0A067QPB4 A0A0N7Z8C7 A0A336KST5 A0A0V0G8I2 A0A023EZJ1 A0A034V9L3 A0A3L8DZ32 A0A224XA48 A0A2J7RCK1 A0A026W464 A0A1B6CDZ4 K7INU7 E0VRI1 A0A0L0BLB6 A0A182NR07 A0A1I8PZD2 A0A182YNB1 A0A0A1X667 A0A1I8MZQ6 A0A182QVW4 A0A240SWJ3 A0A0K8U9N1 A0A182RG15 A0A182VW86 Q7PWZ2 A0A182M491 E9J500 A0A3F2YZN1 A0A182JXI5 A0A182XNY6 A0A182L8B3 A0A1S4GBP6 A0A182HK32 A0A182VC05 A0A1A9W310 W5J5B6 A0A2C9GGW2 A0A151X421 A0A182PR64 A0A1D2N5Q8 A0A195EYS6 A0A195BPC8 A0A158NDR4 A0A195D7M0 F4W842 A0A1A9VU92 A0A1B0B5Y4 A0A0T6ATH7 A0A1B0EYY0 A0A1B0GL72 A0A1B0FM65 D6WCT7 A0A2P8YDJ4 A0A182IRZ8 A0A1Q3EYT2 E2A0N9 A0A1A9ZK21 A0A0A9ZCX4 B4K6N0 A0A0A9Z328 A0A1B6KX76 T1HJ60 A0A1Y1K7Q4 A0A0K8TH28 N6UST1 A0A0Q9WR14 Q176V1 A0A1S4FD04 A0A182GEY8 B4QZX8 B4HE49 A0A1B6FVA1 B3P7M5 A0A3B0JG78 B4JRM7 B4G5B6 I5ANR3 B4PLL3 B4NHS4 A0A0P5L4A4 A0A0B4KGP2 Q9VD44 A0A1W4VPD4
A0A437AXT4 A0A088A209 A0A0L7QUF1 A0A154PRA7 A0A0N0BJF6 A0A067QPB4 A0A0N7Z8C7 A0A336KST5 A0A0V0G8I2 A0A023EZJ1 A0A034V9L3 A0A3L8DZ32 A0A224XA48 A0A2J7RCK1 A0A026W464 A0A1B6CDZ4 K7INU7 E0VRI1 A0A0L0BLB6 A0A182NR07 A0A1I8PZD2 A0A182YNB1 A0A0A1X667 A0A1I8MZQ6 A0A182QVW4 A0A240SWJ3 A0A0K8U9N1 A0A182RG15 A0A182VW86 Q7PWZ2 A0A182M491 E9J500 A0A3F2YZN1 A0A182JXI5 A0A182XNY6 A0A182L8B3 A0A1S4GBP6 A0A182HK32 A0A182VC05 A0A1A9W310 W5J5B6 A0A2C9GGW2 A0A151X421 A0A182PR64 A0A1D2N5Q8 A0A195EYS6 A0A195BPC8 A0A158NDR4 A0A195D7M0 F4W842 A0A1A9VU92 A0A1B0B5Y4 A0A0T6ATH7 A0A1B0EYY0 A0A1B0GL72 A0A1B0FM65 D6WCT7 A0A2P8YDJ4 A0A182IRZ8 A0A1Q3EYT2 E2A0N9 A0A1A9ZK21 A0A0A9ZCX4 B4K6N0 A0A0A9Z328 A0A1B6KX76 T1HJ60 A0A1Y1K7Q4 A0A0K8TH28 N6UST1 A0A0Q9WR14 Q176V1 A0A1S4FD04 A0A182GEY8 B4QZX8 B4HE49 A0A1B6FVA1 B3P7M5 A0A3B0JG78 B4JRM7 B4G5B6 I5ANR3 B4PLL3 B4NHS4 A0A0P5L4A4 A0A0B4KGP2 Q9VD44 A0A1W4VPD4
PDB
4ZRL
E-value=1.20639e-57,
Score=566
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
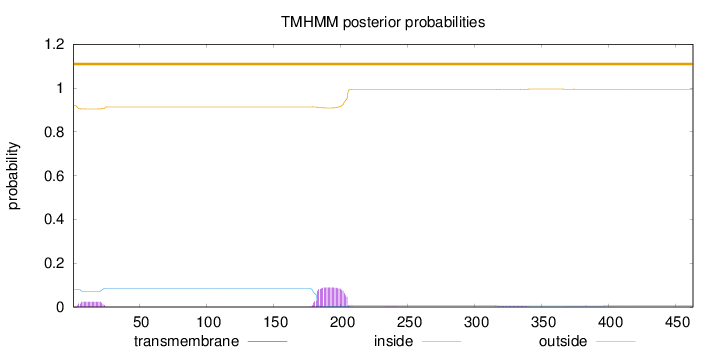
Length:
463
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.46172
Exp number, first 60 AAs:
0.43124
Total prob of N-in:
0.08137
outside
1 - 463
Population Genetic Test Statistics
Pi
272.135617
Theta
165.052425
Tajima's D
1.730896
CLR
0.288891
CSRT
0.834108294585271
Interpretation
Uncertain
