Gene
KWMTBOMO12350 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012388
Annotation
hypothetical_protein_KGM_08900_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 2.438
Sequence
CDS
ATGATGGCTGCAAATAATTATTTTGGGTTCACTCACGGAGGAACCCAATACGGCGCGGCTACAGCTAGTGCTGCATATGGTGGTCAAACCGGGTACGCCGTAGCGCCCGCTGCCACTGCCGCCACTTACGGCACCCAACGAGCCGCTGCCACTGGCTATGATACAGCATATCAAGCTGCGGCTGCCACACAAGCTGCTGCACATGCCCACGCCCACGCAGCAGTAGCAGCGGCCTCAGCATATGACGCCAGCAAGAGTGCGTATTACCAACAGGCAGCTGCTGCCTACCCGACAGCGGCACCCCCACAACCACAACCTACATATGATGCATCTGCAGCTAAACCTGCATACTCCACACCTGCGTCATATGCACAGGGCACATACAGCCAGGGTGCAGCCCAAAGGGGAGCGGCTGCAGCAAAACCTGCTTACGGAGCAGTGTACAGTGCCACCACTGCCACTGCTTCCTACCCCACACAGGCATACACTGCCACCCCGGCACAGACACAGAAACAAACGACGAACAGAGGGAATACCGCCTACGACACCGCGCTCTATAACGCGGCCTCCATGTACGTCGCGCAGCAGAGCAAGGCCGCGCCCGGCACTGGCTGGAAGAACTATAAAACGCCGGGAACGGGTCAGCCTCGCCGCGCCAAGCCTCCGCCCAAAGCGCAACAGTTGCATTACTGTGACGTGTGCCGCATCTCCTGCGCCGGACCCCAGACATACAAGGAGCACCTGGAGGGCCAGAAGCACAAGAAGAAGGAGGCGGCCGTGAAGCTGGCGGCGGCGGGCGGCGGCGCCAGCGCGGCGGGGGGCGCGCGCGGGGCGGCGGGCGGCGCGCTGCGCTGCGAGCTGTGTGACGTCACGTGCACGGGCGCCGACGCGTACGCCGCCCACGTGCGCGGCATCAAGCATCAGAAGGTCGTCAAGCTGCACACCATGCTCGGGAAACCCATCCCCTCCACGGAACCCACTAAAGTCGGCGGAGCCAAAAAGAAAGTGGCGGGCGCGCCGAAGATCGCGTTCGTGTCGTCCGGAGGACTCCGCACTGTCGCCACCGACCCCGCCTCACCCTCCGCGCCCTCCACCGCGCCCTCCACCGCACACTCGACCCCACACTCGCCCCCAGACAAGGAAAAGGACGAGGAGAAGGAGGACGACGATGCCGAGCCCGAGGTGCAGCCCGTGGGACAGGACTACATCGAGGAGATCCGGGCAGACGACGGCAAGGCGCTCTCCTTCAACTGCAAGCTCTGCGATTGCAAGTTCAATGACCCCAATGCTAAGGAAATGCACATGAAGGGCAGAAGGCACCGACTACAGTACAAGAAGAAGGTGCAACCAGATCTAGAGGTTAAGGTCAAACCCTCTATGCACCAGCGTAAGTTGGCGGAGGCTAAAGCTCAACGTATGATGGTGAGAGATGAACTCTGGGCCAGAAGACGCATGCATGATGTATATGGCCAGGGTATTGAGGAAGACGAGCGCATGTATTGGGAGGGGGGAATCGAACCGTGGTGGAGAGGTCCTCATCACCATCATCATCATCATGGCATGGGTATGGGCTACGGTCGCCGGCCGGAGACATCTGACGACCGGCACGTGCTCACCAAGCACGCCGAGATCTATCCGGGAGACGCGCAGCTGCAGCACATCCACCGCGCCGTCGCGCACACCGAGCGCGCCCTCAAGTCGCTCTCCGACGCGCTCGCTGACTCCGCCAGGCAAAAGACTGTGCCGGCTAAAACTAAAGTTGATGACAAGAAAGATGACAAGGCTGATGGCAAAGAAGATGGTCGTGATAATCAGTTGTTCTCATTCCTGGCGGAAGGCGAGCAGGCGGGTTCCACCAGCACGGCCGGCGACGCGCCCCGCGCTCTGAAGGGCGTGATGCGCGTGGGGCTGCTCGCCAAGGGCCTGCTGCTGAAGGGCGACCGCGACGTGCAGCTGGTGGTGCTGTGTCACGACCGCCCCACCGTCACCCTGCTCAAGCGCGTCGCCGCCGACCTGCCCCACCACCTCGCCAAAGTCAAGGGCCCCGGCGACGAGCCCAAGTACAAGGTGGAGCTGCTGCCGGCCGAGGGCGCCGTGCAGGTGTCGGACGCGCACGTGGGCGTGCTCGTCAGCCTCACCTCCGCCGTGGTGCGCGAGCCCGCCGACGGGGGCGACATAAAGCGAGACGACAAGGATGTTCTACCGCGGCAGAAGTGTTTGGACGCGTTGGCCGCCCTTCGGCACGCCAAGTGGTTCCAGGCTAGGGCGGCCACGCTGCAGTCCTGCGTCATCATCATCCGCATCCTGCGCGACCTCTGCAGGCGCATCCCGAACTGGACGCCGCTCAACTCATACGCAATGGAACTGCTGACGGCCGGCGTGGTGCAGTCCGCGGGCGCCGCTCTGTCGCCCGGCGAGGCGCTGCGGAGGCTCATGGAGGCCGTCGCCGGCGGCCTGTTGCTCGAACACGGGCCCGGCCTGCGGGACCCCTGCGAGAAGGAACTCGTTGACGCGCTGGGCAACATGCCGCCGCAGAAACGGGAAGATCTGACGGCCTCAGCGCAACAGTTCTTAAGACAGATTGCTTTCAGGCAAATTCATAAGGTGCTCGACATGGAGCCGCTCCCCAAGGCGAAGCATGCGACGGGCGCCTGGAAGTTCCCGCGCAAGCGCAGGCGCTCCAACACTGACAACGAGCACGACGCGCCCAACGGTGAGGGGAAAATAGTGAAGACAGAGGAAAAATCAGACTGTTCGGACACCAACAAAAACTATCCCGTTCAAGCTTATGTTTTAAACTGCCTAACATCCACTCTTCCTTCACAGCAAATTCAATTTCAGGATTGGCCGGAATTGTTGCAATTGCAGATGGCTGATCCAGGATATGGTACACCAAGCAATATAGATCTTCTACTAGGCGCTGATGTGTATGGTCAGGTTTTGTCAGAAGGCATATTGAGAAATCCACCTGGAACCTTGACAGCTCAAAACACACAGCTGGGATGGATCCTGTCGGGACGAGTTTAG
Protein
MMAANNYFGFTHGGTQYGAATASAAYGGQTGYAVAPAATAATYGTQRAAATGYDTAYQAAAATQAAAHAHAHAAVAAASAYDASKSAYYQQAAAAYPTAAPPQPQPTYDASAAKPAYSTPASYAQGTYSQGAAQRGAAAAKPAYGAVYSATTATASYPTQAYTATPAQTQKQTTNRGNTAYDTALYNAASMYVAQQSKAAPGTGWKNYKTPGTGQPRRAKPPPKAQQLHYCDVCRISCAGPQTYKEHLEGQKHKKKEAAVKLAAAGGGASAAGGARGAAGGALRCELCDVTCTGADAYAAHVRGIKHQKVVKLHTMLGKPIPSTEPTKVGGAKKKVAGAPKIAFVSSGGLRTVATDPASPSAPSTAPSTAHSTPHSPPDKEKDEEKEDDDAEPEVQPVGQDYIEEIRADDGKALSFNCKLCDCKFNDPNAKEMHMKGRRHRLQYKKKVQPDLEVKVKPSMHQRKLAEAKAQRMMVRDELWARRRMHDVYGQGIEEDERMYWEGGIEPWWRGPHHHHHHHGMGMGYGRRPETSDDRHVLTKHAEIYPGDAQLQHIHRAVAHTERALKSLSDALADSARQKTVPAKTKVDDKKDDKADGKEDGRDNQLFSFLAEGEQAGSTSTAGDAPRALKGVMRVGLLAKGLLLKGDRDVQLVVLCHDRPTVTLLKRVAADLPHHLAKVKGPGDEPKYKVELLPAEGAVQVSDAHVGVLVSLTSAVVREPADGGDIKRDDKDVLPRQKCLDALAALRHAKWFQARAATLQSCVIIIRILRDLCRRIPNWTPLNSYAMELLTAGVVQSAGAALSPGEALRRLMEAVAGGLLLEHGPGLRDPCEKELVDALGNMPPQKREDLTASAQQFLRQIAFRQIHKVLDMEPLPKAKHATGAWKFPRKRRRSNTDNEHDAPNGEGKIVKTEEKSDCSDTNKNYPVQAYVLNCLTSTLPSQQIQFQDWPELLQLQMADPGYGTPSNIDLLLGADVYGQVLSEGILRNPPGTLTAQNTQLGWILSGRV
Summary
Uniprot
H9JS78
A0A2H1V1E5
A0A3S2NB99
A0A2A4JVF9
A0A1L8DII7
A0A1L8DI88
+ More
V5GWC7 V5GSF8 V5GKF1 A0A310SJT7 A0A154P0D9 A0A151I5X2 A0A158NX44 F4WQ48 E2ANB4 A0A195ECN5 A0A0J7L0D0 A0A151I788 E2BI21 A0A026VTV8 E9IVX8 B3M621 A0A3L8E4U8 A0A0M9A320 A0A0A1WDC6 A0A232EKP1 B4IX87 B4L0Z6 A0A182Q4U4 U4UMA6 A0A1W4XEZ6 T1PBW7 A0A1Y1MJN7 A0A1I8PE16 A0A1I8PE79 Q86BI3 A0A0M3QVZ2 A0A1I8MAX0 A0A1I8MAW3 A0A088AKW6 A0A1A9WG33 A0A1W4UU64 A0A2A3EQ08 A0A1A9ZBP1 A0A1B0G160 B4HJ18 A0A151WY67 A0A1A9UPF0 B4QLU4 A0A1A9XCG5 A0A1B0B8R3 B4PD32 B3NDS1 B4LGR5 W8BAN1 W8BR74 A0A0K8WLW3 A0A0K8VEU2 B4N6W4 A0A2C9K2A4 A0A087TA70 A0A2C9K2B0 A0A2C9K299 A0A2C9K2B7 R7VAN5 A0A433U0U1 N6U0V0 A0A0K2UC28 A0A1Z5KW70 A0A2R5L615 A0A210QVP5 N6UR05
V5GWC7 V5GSF8 V5GKF1 A0A310SJT7 A0A154P0D9 A0A151I5X2 A0A158NX44 F4WQ48 E2ANB4 A0A195ECN5 A0A0J7L0D0 A0A151I788 E2BI21 A0A026VTV8 E9IVX8 B3M621 A0A3L8E4U8 A0A0M9A320 A0A0A1WDC6 A0A232EKP1 B4IX87 B4L0Z6 A0A182Q4U4 U4UMA6 A0A1W4XEZ6 T1PBW7 A0A1Y1MJN7 A0A1I8PE16 A0A1I8PE79 Q86BI3 A0A0M3QVZ2 A0A1I8MAX0 A0A1I8MAW3 A0A088AKW6 A0A1A9WG33 A0A1W4UU64 A0A2A3EQ08 A0A1A9ZBP1 A0A1B0G160 B4HJ18 A0A151WY67 A0A1A9UPF0 B4QLU4 A0A1A9XCG5 A0A1B0B8R3 B4PD32 B3NDS1 B4LGR5 W8BAN1 W8BR74 A0A0K8WLW3 A0A0K8VEU2 B4N6W4 A0A2C9K2A4 A0A087TA70 A0A2C9K2B0 A0A2C9K299 A0A2C9K2B7 R7VAN5 A0A433U0U1 N6U0V0 A0A0K2UC28 A0A1Z5KW70 A0A2R5L615 A0A210QVP5 N6UR05
Pubmed
EMBL
BABH01033092
BABH01033093
ODYU01000225
SOQ34639.1
RSAL01000284
RVE42991.1
+ More
NWSH01000557 PCG75678.1 GFDF01007907 JAV06177.1 GFDF01007896 JAV06188.1 GALX01003928 JAB64538.1 GALX01003929 JAB64537.1 GALX01003927 JAB64539.1 KQ764626 OAD54466.1 KQ434791 KZC05389.1 KQ976434 KYM87213.1 ADTU01002998 GL888262 EGI63671.1 GL441177 EFN65062.1 KQ979074 KYN22871.1 LBMM01001592 KMQ96048.1 KQ978464 KYM93762.1 GL448433 EFN84647.1 KK107921 EZA47228.1 GL766449 EFZ15257.1 CH902618 EDV40737.1 KPU78739.1 KPU78740.1 QOIP01000001 RLU27552.1 KQ435750 KOX76260.1 GBXI01017425 JAC96866.1 NNAY01003733 OXU18929.1 CH916366 EDV97419.1 CH933809 EDW18153.1 AXCN02000191 KB632275 ERL91140.1 KA646261 AFP60890.1 GEZM01029729 JAV85962.1 AE014296 AAO41247.1 AGB94617.1 AGB94618.1 AGB94619.1 CP012525 ALC43243.1 KZ288195 PBC33770.1 CCAG010020780 CCAG010020781 CH480815 EDW41674.1 KQ982651 KYQ52850.1 CM000363 CM002912 EDX10643.1 KMY99937.1 JXJN01010041 CM000159 EDW94964.2 KRK02164.1 CH954178 EDV52204.1 KQS44182.1 KQS44183.1 CH940647 EDW70530.1 KRF84959.1 GAMC01010818 JAB95737.1 GAMC01010819 JAB95736.1 GDHF01000183 JAI52131.1 GDHF01015254 JAI37060.1 CH964168 EDW80103.1 KK114248 KFM62009.1 AMQN01005144 KB295236 ELU13401.1 RQTK01000110 RUS87461.1 APGK01053960 APGK01053961 KB741239 ENN72172.1 HACA01018433 CDW35794.1 GFJQ02007614 JAV99355.1 GGLE01000764 MBY04890.1 NEDP02001616 OWF52827.1 APGK01020224 APGK01020225 APGK01020226 KB740181 ENN81182.1
NWSH01000557 PCG75678.1 GFDF01007907 JAV06177.1 GFDF01007896 JAV06188.1 GALX01003928 JAB64538.1 GALX01003929 JAB64537.1 GALX01003927 JAB64539.1 KQ764626 OAD54466.1 KQ434791 KZC05389.1 KQ976434 KYM87213.1 ADTU01002998 GL888262 EGI63671.1 GL441177 EFN65062.1 KQ979074 KYN22871.1 LBMM01001592 KMQ96048.1 KQ978464 KYM93762.1 GL448433 EFN84647.1 KK107921 EZA47228.1 GL766449 EFZ15257.1 CH902618 EDV40737.1 KPU78739.1 KPU78740.1 QOIP01000001 RLU27552.1 KQ435750 KOX76260.1 GBXI01017425 JAC96866.1 NNAY01003733 OXU18929.1 CH916366 EDV97419.1 CH933809 EDW18153.1 AXCN02000191 KB632275 ERL91140.1 KA646261 AFP60890.1 GEZM01029729 JAV85962.1 AE014296 AAO41247.1 AGB94617.1 AGB94618.1 AGB94619.1 CP012525 ALC43243.1 KZ288195 PBC33770.1 CCAG010020780 CCAG010020781 CH480815 EDW41674.1 KQ982651 KYQ52850.1 CM000363 CM002912 EDX10643.1 KMY99937.1 JXJN01010041 CM000159 EDW94964.2 KRK02164.1 CH954178 EDV52204.1 KQS44182.1 KQS44183.1 CH940647 EDW70530.1 KRF84959.1 GAMC01010818 JAB95737.1 GAMC01010819 JAB95736.1 GDHF01000183 JAI52131.1 GDHF01015254 JAI37060.1 CH964168 EDW80103.1 KK114248 KFM62009.1 AMQN01005144 KB295236 ELU13401.1 RQTK01000110 RUS87461.1 APGK01053960 APGK01053961 KB741239 ENN72172.1 HACA01018433 CDW35794.1 GFJQ02007614 JAV99355.1 GGLE01000764 MBY04890.1 NEDP02001616 OWF52827.1 APGK01020224 APGK01020225 APGK01020226 KB740181 ENN81182.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000076502
UP000078540
UP000005205
+ More
UP000007755 UP000000311 UP000078492 UP000036403 UP000078542 UP000008237 UP000053097 UP000007801 UP000279307 UP000053105 UP000215335 UP000001070 UP000009192 UP000075886 UP000030742 UP000192223 UP000095300 UP000000803 UP000092553 UP000095301 UP000005203 UP000091820 UP000192221 UP000242457 UP000092445 UP000092444 UP000001292 UP000075809 UP000078200 UP000000304 UP000092443 UP000092460 UP000002282 UP000008711 UP000008792 UP000007798 UP000076420 UP000054359 UP000014760 UP000271974 UP000019118 UP000242188
UP000007755 UP000000311 UP000078492 UP000036403 UP000078542 UP000008237 UP000053097 UP000007801 UP000279307 UP000053105 UP000215335 UP000001070 UP000009192 UP000075886 UP000030742 UP000192223 UP000095300 UP000000803 UP000092553 UP000095301 UP000005203 UP000091820 UP000192221 UP000242457 UP000092445 UP000092444 UP000001292 UP000075809 UP000078200 UP000000304 UP000092443 UP000092460 UP000002282 UP000008711 UP000008792 UP000007798 UP000076420 UP000054359 UP000014760 UP000271974 UP000019118 UP000242188
Interpro
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
H9JS78
A0A2H1V1E5
A0A3S2NB99
A0A2A4JVF9
A0A1L8DII7
A0A1L8DI88
+ More
V5GWC7 V5GSF8 V5GKF1 A0A310SJT7 A0A154P0D9 A0A151I5X2 A0A158NX44 F4WQ48 E2ANB4 A0A195ECN5 A0A0J7L0D0 A0A151I788 E2BI21 A0A026VTV8 E9IVX8 B3M621 A0A3L8E4U8 A0A0M9A320 A0A0A1WDC6 A0A232EKP1 B4IX87 B4L0Z6 A0A182Q4U4 U4UMA6 A0A1W4XEZ6 T1PBW7 A0A1Y1MJN7 A0A1I8PE16 A0A1I8PE79 Q86BI3 A0A0M3QVZ2 A0A1I8MAX0 A0A1I8MAW3 A0A088AKW6 A0A1A9WG33 A0A1W4UU64 A0A2A3EQ08 A0A1A9ZBP1 A0A1B0G160 B4HJ18 A0A151WY67 A0A1A9UPF0 B4QLU4 A0A1A9XCG5 A0A1B0B8R3 B4PD32 B3NDS1 B4LGR5 W8BAN1 W8BR74 A0A0K8WLW3 A0A0K8VEU2 B4N6W4 A0A2C9K2A4 A0A087TA70 A0A2C9K2B0 A0A2C9K299 A0A2C9K2B7 R7VAN5 A0A433U0U1 N6U0V0 A0A0K2UC28 A0A1Z5KW70 A0A2R5L615 A0A210QVP5 N6UR05
V5GWC7 V5GSF8 V5GKF1 A0A310SJT7 A0A154P0D9 A0A151I5X2 A0A158NX44 F4WQ48 E2ANB4 A0A195ECN5 A0A0J7L0D0 A0A151I788 E2BI21 A0A026VTV8 E9IVX8 B3M621 A0A3L8E4U8 A0A0M9A320 A0A0A1WDC6 A0A232EKP1 B4IX87 B4L0Z6 A0A182Q4U4 U4UMA6 A0A1W4XEZ6 T1PBW7 A0A1Y1MJN7 A0A1I8PE16 A0A1I8PE79 Q86BI3 A0A0M3QVZ2 A0A1I8MAX0 A0A1I8MAW3 A0A088AKW6 A0A1A9WG33 A0A1W4UU64 A0A2A3EQ08 A0A1A9ZBP1 A0A1B0G160 B4HJ18 A0A151WY67 A0A1A9UPF0 B4QLU4 A0A1A9XCG5 A0A1B0B8R3 B4PD32 B3NDS1 B4LGR5 W8BAN1 W8BR74 A0A0K8WLW3 A0A0K8VEU2 B4N6W4 A0A2C9K2A4 A0A087TA70 A0A2C9K2B0 A0A2C9K299 A0A2C9K2B7 R7VAN5 A0A433U0U1 N6U0V0 A0A0K2UC28 A0A1Z5KW70 A0A2R5L615 A0A210QVP5 N6UR05
PDB
4ATB
E-value=3.60916e-54,
Score=539
Ontologies
GO
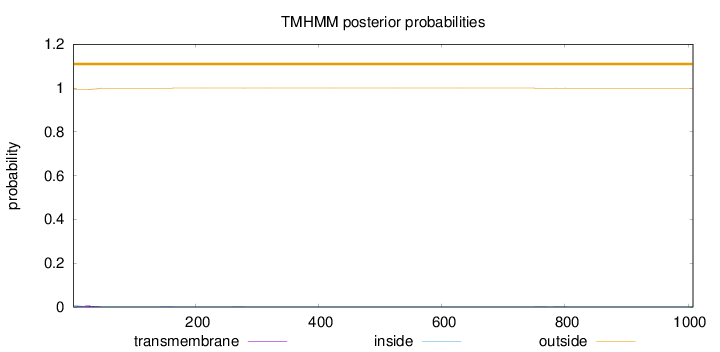
Topology
Length:
1008
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.186860000000001
Exp number, first 60 AAs:
0.16
Total prob of N-in:
0.00753
outside
1 - 1008
Population Genetic Test Statistics
Pi
194.102507
Theta
178.167371
Tajima's D
-1.438814
CLR
0.442262
CSRT
0.0686965651717414
Interpretation
Uncertain
