Gene
KWMTBOMO12349 Validated by peptides from experiments
Annotation
PREDICTED:_immediate_early_response_3-interacting_protein_1-like_[Plutella_xylostella]
Full name
Immediate early response 3-interacting protein 1
Location in the cell
Extracellular Reliability : 2.16
Sequence
CDS
ATGCTCACTTTATGGAATTTATTTGAAGCAACATTGTTATGTCTCAACGCAGTATGTGTATTACACGAAGAAAGATTCATGCAGAAAATTGGCTGGGGAGCAAATACCCAAACCCAGGGCTTTGAAGATCAATCTTCGATGAAATTCCAAATTCTTAACTTGGTGAGGTCAATTAGAACAGTTACAAAGATTCCTCTCATCATACTCAATATCCTGACAATAATGTTCAAACTGTTACTGGGATAA
Protein
MLTLWNLFEATLLCLNAVCVLHEERFMQKIGWGANTQTQGFEDQSSMKFQILNLVRSIRTVTKIPLIILNILTIMFKLLLG
Summary
Description
May be involved in protein transport between endoplasmic reticulum and Golgi apparatus.
May be implicated in the regulation of apoptosis. May be involved in protein transport between endoplasmic reticulum and Golgi apparatus.
May be implicated in the regulation of apoptosis. May be involved in protein transport between endoplasmic reticulum and Golgi apparatus.
Similarity
Belongs to the YOS1 family.
Keywords
Endoplasmic reticulum
Membrane
Transmembrane
Transmembrane helix
Complete proteome
Reference proteome
Feature
chain Immediate early response 3-interacting protein 1
Uniprot
A0A212FBC2
A0A3S2NRY1
A0A2W1BYQ9
A0A2P8Y5F8
S4Q0C5
U5EU61
+ More
A0A194RA40 I4DLK9 A0A2H1V1B8 A0A023EDD3 Q16NK9 A0A194PWB8 A0A1B0CG41 A0A1L8DD25 A0A1Q3FHU0 B0WY61 A0A182K6Y3 A0A1L8EEC7 A0A182UNW9 A0A453YZF5 A0A1I8P3F6 A0A182N7W0 T1IWQ8 A0A182FGT4 W5JTL3 A0A1I8MPV0 A0A0L0C103 A0A2M4C579 A0A2M4ASS0 D2A526 A0A0T6AVY7 A0A2M4AR77 A0A1B0GGC4 A0A1A9ZWL9 E2C9U0 K7INU8 B4LEG9 B4KWF3 B4J1Q1 N6TLE9 B4N438 A0A0L8H2E4 A0A2M3ZC78 A0A0M8ZQW4 Q2LZZ2 B4H932 B3M5N6 A0A1W4W4N5 A0A0M5J3W7 E9IAE4 A0A154PGH7 A0A195AUV7 A0A158P0Y6 A0A026VXR6 B4PEK1 B4HDU6 Q9VTE1 A0A151ILU8 A0A151JXY9 B4QPN1 A0A2A3EAM8 A0A087ZMW2 B3NCQ9 T1D6K8 E1ZW58 A0A0L7R731 A0A2P8Y5F5 A0A087U5V6 A0A0K8RLZ7 A0A151J1M4 K1Q9Z0 A0A023GHI6 G3MTU5 V4AZX4 T1G066 A0A433SL51 F4WTX1 A0A1J1I920 A0A1B0GP98 A0A0N8C8B2 A0A1D2MZB5 Q3B8G7 A0A224YRI4 A0A131Z4W4 A0A131XPU7 A0A182G3J3 A0A0P6C8N0 W4XCI6 A0A2J7Q365 A0A341AIM8 A0A3Q7Q9H4 A0A2Y9GR63 A0A2U3YV64 A0A3Q7SAV5 A0A2Y9ISW8 A0A452S3I5 A0A3Q7WC86 A0A384C4V4 P85007 A0A067QPC3
A0A194RA40 I4DLK9 A0A2H1V1B8 A0A023EDD3 Q16NK9 A0A194PWB8 A0A1B0CG41 A0A1L8DD25 A0A1Q3FHU0 B0WY61 A0A182K6Y3 A0A1L8EEC7 A0A182UNW9 A0A453YZF5 A0A1I8P3F6 A0A182N7W0 T1IWQ8 A0A182FGT4 W5JTL3 A0A1I8MPV0 A0A0L0C103 A0A2M4C579 A0A2M4ASS0 D2A526 A0A0T6AVY7 A0A2M4AR77 A0A1B0GGC4 A0A1A9ZWL9 E2C9U0 K7INU8 B4LEG9 B4KWF3 B4J1Q1 N6TLE9 B4N438 A0A0L8H2E4 A0A2M3ZC78 A0A0M8ZQW4 Q2LZZ2 B4H932 B3M5N6 A0A1W4W4N5 A0A0M5J3W7 E9IAE4 A0A154PGH7 A0A195AUV7 A0A158P0Y6 A0A026VXR6 B4PEK1 B4HDU6 Q9VTE1 A0A151ILU8 A0A151JXY9 B4QPN1 A0A2A3EAM8 A0A087ZMW2 B3NCQ9 T1D6K8 E1ZW58 A0A0L7R731 A0A2P8Y5F5 A0A087U5V6 A0A0K8RLZ7 A0A151J1M4 K1Q9Z0 A0A023GHI6 G3MTU5 V4AZX4 T1G066 A0A433SL51 F4WTX1 A0A1J1I920 A0A1B0GP98 A0A0N8C8B2 A0A1D2MZB5 Q3B8G7 A0A224YRI4 A0A131Z4W4 A0A131XPU7 A0A182G3J3 A0A0P6C8N0 W4XCI6 A0A2J7Q365 A0A341AIM8 A0A3Q7Q9H4 A0A2Y9GR63 A0A2U3YV64 A0A3Q7SAV5 A0A2Y9ISW8 A0A452S3I5 A0A3Q7WC86 A0A384C4V4 P85007 A0A067QPC3
Pubmed
22118469
28756777
29403074
23622113
26354079
22651552
+ More
24945155 17510324 12364791 20920257 23761445 25315136 26108605 18362917 19820115 20798317 20075255 17994087 18057021 23537049 15632085 21282665 21347285 24508170 30249741 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 24330624 22992520 22216098 23254933 21719571 27289101 28797301 26830274 28049606 26483478 24813606 15057822 24845553
24945155 17510324 12364791 20920257 23761445 25315136 26108605 18362917 19820115 20798317 20075255 17994087 18057021 23537049 15632085 21282665 21347285 24508170 30249741 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 24330624 22992520 22216098 23254933 21719571 27289101 28797301 26830274 28049606 26483478 24813606 15057822 24845553
EMBL
AGBW02009347
OWR51042.1
RSAL01000284
RVE42992.1
KZ149941
PZC76913.1
+ More
PYGN01000906 PSN39489.1 GAIX01000028 JAA92532.1 GANO01001634 JAB58237.1 KQ460473 KPJ14492.1 AK402177 BAM18799.1 ODYU01000225 SOQ34638.1 GAPW01006737 JAC06861.1 CH477819 CH477231 EAT35938.1 EAT46900.1 KQ459591 KPI97054.1 AJWK01010713 GFDF01009799 JAV04285.1 GFDL01007915 JAV27130.1 DS232182 EDS36938.1 GFDG01001866 JAV16933.1 AAAB01008960 JH431628 ADMH02000528 ETN66089.1 JRES01001060 KNC25931.1 GGFJ01011278 MBW60419.1 GGFK01010502 MBW43823.1 KQ971361 EFA05304.2 LJIG01022671 KRT79327.1 GGFK01009958 MBW43279.1 CCAG010015091 GL453902 EFN75277.1 CH940647 EDW70145.1 KRF84736.1 CH933809 EDW19582.1 CH916366 EDV96971.1 APGK01019842 KB740134 KB632275 ENN81279.1 ERL91086.1 CH964095 EDW78912.1 KQ419500 KOF83392.1 GGFM01005361 MBW26112.1 KQ435896 KOX69285.1 CH379069 EAL31145.2 CH479226 EDW35247.1 CH902618 EDV40670.1 CP012525 ALC44281.1 GL762003 EFZ22475.1 KQ434890 KZC10418.1 KQ976738 KYM75759.1 ADTU01005902 KK107602 QOIP01000001 EZA48562.1 RLU26605.1 RLU26683.1 CM000159 EDW94067.1 CH480815 EDW41036.1 AE014296 BT023689 KX531733 AAF50111.2 AAY85089.1 ANY27543.1 KQ977086 KYN05865.1 KQ981515 KYN40764.1 CM000363 CM002912 EDX10022.1 KMY98911.1 KZ288302 PBC28823.1 CH954178 EDV51356.1 GALA01000013 JAA94839.1 GL434776 EFN74567.1 KQ414646 KOC66561.1 PSN39488.1 KK118333 KFM72745.1 GADI01001691 JAA72117.1 KQ980514 KYN15742.1 JH823254 EKC18276.1 GBBM01003218 JAC32200.1 JO845297 AEO36913.1 KB200049 ESP03298.1 AMQM01002059 KB097700 ESN91416.1 RQTK01001555 RUS69854.1 GL888345 EGI62328.1 CVRI01000042 CRK95462.1 AJVK01015029 GDIQ01104963 LRGB01001005 JAL46763.1 KZS14028.1 LJIJ01000360 ODM98383.1 BC106448 AAI06449.1 GFPF01009071 MAA20217.1 GEDV01002108 JAP86449.1 GEFH01000356 JAP68225.1 JXUM01040886 KQ561263 KXJ79165.1 GDIP01011509 JAM92206.1 AAGJ04069297 NEVH01019070 PNF23010.1 AABR03109891 KK853171 KDR10325.1
PYGN01000906 PSN39489.1 GAIX01000028 JAA92532.1 GANO01001634 JAB58237.1 KQ460473 KPJ14492.1 AK402177 BAM18799.1 ODYU01000225 SOQ34638.1 GAPW01006737 JAC06861.1 CH477819 CH477231 EAT35938.1 EAT46900.1 KQ459591 KPI97054.1 AJWK01010713 GFDF01009799 JAV04285.1 GFDL01007915 JAV27130.1 DS232182 EDS36938.1 GFDG01001866 JAV16933.1 AAAB01008960 JH431628 ADMH02000528 ETN66089.1 JRES01001060 KNC25931.1 GGFJ01011278 MBW60419.1 GGFK01010502 MBW43823.1 KQ971361 EFA05304.2 LJIG01022671 KRT79327.1 GGFK01009958 MBW43279.1 CCAG010015091 GL453902 EFN75277.1 CH940647 EDW70145.1 KRF84736.1 CH933809 EDW19582.1 CH916366 EDV96971.1 APGK01019842 KB740134 KB632275 ENN81279.1 ERL91086.1 CH964095 EDW78912.1 KQ419500 KOF83392.1 GGFM01005361 MBW26112.1 KQ435896 KOX69285.1 CH379069 EAL31145.2 CH479226 EDW35247.1 CH902618 EDV40670.1 CP012525 ALC44281.1 GL762003 EFZ22475.1 KQ434890 KZC10418.1 KQ976738 KYM75759.1 ADTU01005902 KK107602 QOIP01000001 EZA48562.1 RLU26605.1 RLU26683.1 CM000159 EDW94067.1 CH480815 EDW41036.1 AE014296 BT023689 KX531733 AAF50111.2 AAY85089.1 ANY27543.1 KQ977086 KYN05865.1 KQ981515 KYN40764.1 CM000363 CM002912 EDX10022.1 KMY98911.1 KZ288302 PBC28823.1 CH954178 EDV51356.1 GALA01000013 JAA94839.1 GL434776 EFN74567.1 KQ414646 KOC66561.1 PSN39488.1 KK118333 KFM72745.1 GADI01001691 JAA72117.1 KQ980514 KYN15742.1 JH823254 EKC18276.1 GBBM01003218 JAC32200.1 JO845297 AEO36913.1 KB200049 ESP03298.1 AMQM01002059 KB097700 ESN91416.1 RQTK01001555 RUS69854.1 GL888345 EGI62328.1 CVRI01000042 CRK95462.1 AJVK01015029 GDIQ01104963 LRGB01001005 JAL46763.1 KZS14028.1 LJIJ01000360 ODM98383.1 BC106448 AAI06449.1 GFPF01009071 MAA20217.1 GEDV01002108 JAP86449.1 GEFH01000356 JAP68225.1 JXUM01040886 KQ561263 KXJ79165.1 GDIP01011509 JAM92206.1 AAGJ04069297 NEVH01019070 PNF23010.1 AABR03109891 KK853171 KDR10325.1
Proteomes
UP000007151
UP000283053
UP000245037
UP000053240
UP000008820
UP000053268
+ More
UP000092461 UP000002320 UP000075881 UP000075903 UP000095300 UP000075884 UP000069272 UP000000673 UP000095301 UP000037069 UP000007266 UP000092444 UP000092445 UP000008237 UP000002358 UP000008792 UP000009192 UP000001070 UP000019118 UP000030742 UP000007798 UP000053454 UP000053105 UP000001819 UP000008744 UP000007801 UP000192221 UP000092553 UP000076502 UP000078540 UP000005205 UP000053097 UP000279307 UP000002282 UP000001292 UP000000803 UP000078542 UP000078541 UP000000304 UP000242457 UP000005203 UP000008711 UP000000311 UP000053825 UP000054359 UP000078492 UP000005408 UP000030746 UP000015101 UP000271974 UP000007755 UP000183832 UP000092462 UP000076858 UP000094527 UP000069940 UP000249989 UP000007110 UP000235965 UP000252040 UP000286641 UP000248481 UP000245341 UP000286640 UP000248482 UP000291022 UP000286642 UP000261680 UP000291021 UP000002494 UP000027135
UP000092461 UP000002320 UP000075881 UP000075903 UP000095300 UP000075884 UP000069272 UP000000673 UP000095301 UP000037069 UP000007266 UP000092444 UP000092445 UP000008237 UP000002358 UP000008792 UP000009192 UP000001070 UP000019118 UP000030742 UP000007798 UP000053454 UP000053105 UP000001819 UP000008744 UP000007801 UP000192221 UP000092553 UP000076502 UP000078540 UP000005205 UP000053097 UP000279307 UP000002282 UP000001292 UP000000803 UP000078542 UP000078541 UP000000304 UP000242457 UP000005203 UP000008711 UP000000311 UP000053825 UP000054359 UP000078492 UP000005408 UP000030746 UP000015101 UP000271974 UP000007755 UP000183832 UP000092462 UP000076858 UP000094527 UP000069940 UP000249989 UP000007110 UP000235965 UP000252040 UP000286641 UP000248481 UP000245341 UP000286640 UP000248482 UP000291022 UP000286642 UP000261680 UP000291021 UP000002494 UP000027135
Interpro
Gene 3D
ProteinModelPortal
A0A212FBC2
A0A3S2NRY1
A0A2W1BYQ9
A0A2P8Y5F8
S4Q0C5
U5EU61
+ More
A0A194RA40 I4DLK9 A0A2H1V1B8 A0A023EDD3 Q16NK9 A0A194PWB8 A0A1B0CG41 A0A1L8DD25 A0A1Q3FHU0 B0WY61 A0A182K6Y3 A0A1L8EEC7 A0A182UNW9 A0A453YZF5 A0A1I8P3F6 A0A182N7W0 T1IWQ8 A0A182FGT4 W5JTL3 A0A1I8MPV0 A0A0L0C103 A0A2M4C579 A0A2M4ASS0 D2A526 A0A0T6AVY7 A0A2M4AR77 A0A1B0GGC4 A0A1A9ZWL9 E2C9U0 K7INU8 B4LEG9 B4KWF3 B4J1Q1 N6TLE9 B4N438 A0A0L8H2E4 A0A2M3ZC78 A0A0M8ZQW4 Q2LZZ2 B4H932 B3M5N6 A0A1W4W4N5 A0A0M5J3W7 E9IAE4 A0A154PGH7 A0A195AUV7 A0A158P0Y6 A0A026VXR6 B4PEK1 B4HDU6 Q9VTE1 A0A151ILU8 A0A151JXY9 B4QPN1 A0A2A3EAM8 A0A087ZMW2 B3NCQ9 T1D6K8 E1ZW58 A0A0L7R731 A0A2P8Y5F5 A0A087U5V6 A0A0K8RLZ7 A0A151J1M4 K1Q9Z0 A0A023GHI6 G3MTU5 V4AZX4 T1G066 A0A433SL51 F4WTX1 A0A1J1I920 A0A1B0GP98 A0A0N8C8B2 A0A1D2MZB5 Q3B8G7 A0A224YRI4 A0A131Z4W4 A0A131XPU7 A0A182G3J3 A0A0P6C8N0 W4XCI6 A0A2J7Q365 A0A341AIM8 A0A3Q7Q9H4 A0A2Y9GR63 A0A2U3YV64 A0A3Q7SAV5 A0A2Y9ISW8 A0A452S3I5 A0A3Q7WC86 A0A384C4V4 P85007 A0A067QPC3
A0A194RA40 I4DLK9 A0A2H1V1B8 A0A023EDD3 Q16NK9 A0A194PWB8 A0A1B0CG41 A0A1L8DD25 A0A1Q3FHU0 B0WY61 A0A182K6Y3 A0A1L8EEC7 A0A182UNW9 A0A453YZF5 A0A1I8P3F6 A0A182N7W0 T1IWQ8 A0A182FGT4 W5JTL3 A0A1I8MPV0 A0A0L0C103 A0A2M4C579 A0A2M4ASS0 D2A526 A0A0T6AVY7 A0A2M4AR77 A0A1B0GGC4 A0A1A9ZWL9 E2C9U0 K7INU8 B4LEG9 B4KWF3 B4J1Q1 N6TLE9 B4N438 A0A0L8H2E4 A0A2M3ZC78 A0A0M8ZQW4 Q2LZZ2 B4H932 B3M5N6 A0A1W4W4N5 A0A0M5J3W7 E9IAE4 A0A154PGH7 A0A195AUV7 A0A158P0Y6 A0A026VXR6 B4PEK1 B4HDU6 Q9VTE1 A0A151ILU8 A0A151JXY9 B4QPN1 A0A2A3EAM8 A0A087ZMW2 B3NCQ9 T1D6K8 E1ZW58 A0A0L7R731 A0A2P8Y5F5 A0A087U5V6 A0A0K8RLZ7 A0A151J1M4 K1Q9Z0 A0A023GHI6 G3MTU5 V4AZX4 T1G066 A0A433SL51 F4WTX1 A0A1J1I920 A0A1B0GP98 A0A0N8C8B2 A0A1D2MZB5 Q3B8G7 A0A224YRI4 A0A131Z4W4 A0A131XPU7 A0A182G3J3 A0A0P6C8N0 W4XCI6 A0A2J7Q365 A0A341AIM8 A0A3Q7Q9H4 A0A2Y9GR63 A0A2U3YV64 A0A3Q7SAV5 A0A2Y9ISW8 A0A452S3I5 A0A3Q7WC86 A0A384C4V4 P85007 A0A067QPC3
Ontologies
PANTHER
Topology
Subcellular location
Endoplasmic reticulum membrane
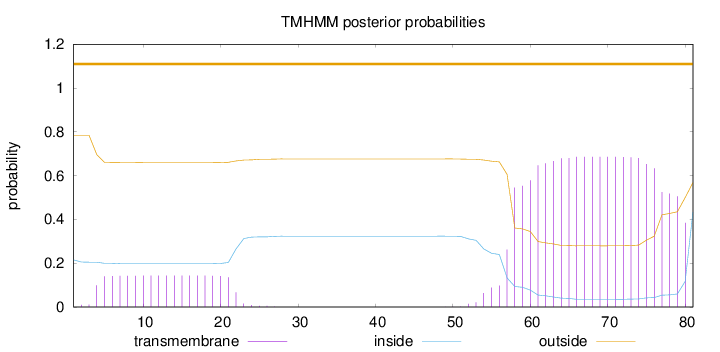
Length:
81
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
17.57692
Exp number, first 60 AAs:
4.86744
Total prob of N-in:
0.21539
outside
1 - 81
Population Genetic Test Statistics
Pi
77.575742
Theta
136.054839
Tajima's D
-0.510599
CLR
0.156087
CSRT
0.24348782560872
Interpretation
Uncertain
