Gene
KWMTBOMO12347 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000209
Annotation
PREDICTED:_fibrillin-1-like_[Bombyx_mori]
Full name
Fibrillin-2
+ More
Fibrillin-1
Fibrillin-3
Latent-transforming growth factor beta-binding protein 2
Latent-transforming growth factor beta-binding protein 1
Latent-transforming growth factor beta-binding protein 4
Latent-transforming growth factor beta-binding protein 3
Neurogenic locus notch homolog protein 1
Neurogenic locus notch homolog protein 2
Neurogenic locus notch homolog protein 3
Neurogenic locus Notch protein
Fibropellin-1
Protein eyes shut homolog
Multiple epidermal growth factor-like domains protein 6
Neurogenic locus notch homolog protein 4
Adhesion G protein-coupled receptor E2
Protein crumbs
Protein jagged-1
Nephronectin
Fibrillin-1
Fibrillin-3
Latent-transforming growth factor beta-binding protein 2
Latent-transforming growth factor beta-binding protein 1
Latent-transforming growth factor beta-binding protein 4
Latent-transforming growth factor beta-binding protein 3
Neurogenic locus notch homolog protein 1
Neurogenic locus notch homolog protein 2
Neurogenic locus notch homolog protein 3
Neurogenic locus Notch protein
Fibropellin-1
Protein eyes shut homolog
Multiple epidermal growth factor-like domains protein 6
Neurogenic locus notch homolog protein 4
Adhesion G protein-coupled receptor E2
Protein crumbs
Protein jagged-1
Nephronectin
Alternative Name
MP340
Transforming growth factor beta-1-binding protein 1
Transforming growth factor beta-1-masking protein large subunit
Motch A
mT14
p300
Translocation-associated notch protein TAN-1
Motch B
Epidermal growth factor-related protein 1
Fibropellin-I
SpEGF I
UEGF-1
Epidermal growth factor-like protein 3
EGF-like module receptor 2
EGF-like module-containing mucin-like hormone receptor-like 2
95F
Preosteoblast EGF-like repeat protein with MAM domain
Protein EGFL6-like
Transforming growth factor beta-1-binding protein 1
Transforming growth factor beta-1-masking protein large subunit
Motch A
mT14
p300
Translocation-associated notch protein TAN-1
Motch B
Epidermal growth factor-related protein 1
Fibropellin-I
SpEGF I
UEGF-1
Epidermal growth factor-like protein 3
EGF-like module receptor 2
EGF-like module-containing mucin-like hormone receptor-like 2
95F
Preosteoblast EGF-like repeat protein with MAM domain
Protein EGFL6-like
Location in the cell
Extracellular Reliability : 3.055
Sequence
CDS
ATGAGTTATTGGCGGTTGTTAGTGGCGGCGGCGCTGCTGGCCAGCGTGGCGGGAGACCCTCGCACGCCGCGCGGCACCACGCGGCCGCGCCACTGGCAGCGAGGACAACGCATGCTCGACGATCGAGCGCCCAGCCAGTTTGTCGGTTCACACGTATGCCGGAGCCGTAACAGCACGCATTGCTGTCCCGGGTGGACATCCAGACCTAATTCATTGCTGTGTATCGTCCCACTATGCCGACCAGACTGCGGCTCGCCAGGTGCTTGTATAGCTCCGAACATCTGTCGATGTCCAGGAGGAGTCGAGGCGCCTTCCTGTGGGAATGGCGGTTTATACCCTTATCCAGTGAGGACACAAGGCGGTTGTCGACGTATCTGTATGAACGGGGGCACGTGTACGAACGGCACATGCGCCTGTGCTCCCGGCTGGAGCGGGGAGTTCTGCACAGAACCGATATGTAAGGAGCCATGTTTGCACGGCGGCCGCTGTATTGCGCCGGACAGATGCGTGTGTTTCCACGGTCTCTCCGGTCCGAGATGCGAGATCGATAGGAGAACAGGTCCGTGCTACACGGAGACGCGTGGCGCGCTGTGCACGGGCGCGCTGGAGGGCGTGGTATGTACGCGGCAGCTCTGCTGTGCCACGGTCGGCCGCGCCTGGGGACACCCCTGCGAGAGGTGCTCCGACCTCGACTGTCCAGTGGGACACCTGCGGAACCTCGCTACCAAGGAGTGCCAGGACATAGACGAGTGCGCAGCTGTACCGGGCTTGTGTTCAGGGGGCCGTTGCATCAACTCCATAGGGTCATTCTCATGCGAATGTCCGCCAGGACAGCGCCGGCATCCCCTCTCCAACAACTGCGAAGACATCGACGAATGCGAGGATCCCGATATTTGTCCTAATGGCAAATGTGTGAACACGGAAGGAGATTACTACTGTCTTTGCAACCCCCACTTCATTCCTAGTCCTGACAAGAAATTCTGTATTGACGGGCGAGTGGGCAGCTGCTACGCGTACCTGAGCGAGACGGGTGAGTGCGCGGACCGGCTGCCGGTGGCGCTGTCCAACCGCGACTGCTGCTGCGGGTACAACATGGGCCGCGCCTGGGGCGACCTCTGCACGCCGTGCCCGCCGCGCGGCGCCACTGAATGGACGCATCTGTGTGGTGGTGGTGCTATCCCTCCGAACTGGAGGAGGAACGTGACAGGAGGAGGTGGAGGTGGTGGCATTGGTGGGGGAGGACAGGGTGGGGAAGGGGGAGGCTCATCTGGAGGCGGAGAAGATGGTGACGGCGGAGGAGGAACATTCGCGCCTCCATCCATAGCGAAGATCAACGAGTGCATGCTAAGAAACGGTATCTGCGGATTGGGAAATTGTATTGATAAGGATGTCGGGTACACCTGCGAGTGCTGGGCCGGCGCGGAGGTGGCAGAGATCGACGGGAACCGCGTCTGCATCGACATCGACGAGTGCGCCCTCGAGTACTGCAAGGGGGGGACGTGCATCAACCGACCGGGGGACTTCGAGTGCAGGTGTCCTCCGGGCTTCGACCCCGTTGAAAACGGTTTGAGATGCAGCGACAAGAACGAATGTGAGATGACCAACGGAGGAATGTGCACCAACGGAATCTGCAAGAATATAGATGGAGGGTTCGAGTGCATCTGCAAGCCCGGGTACGAGTCGGGCTCGGAGGGCGCGCTGTGCCGCGACGTGGACGAGTGCCGCGACAACCCGCGCGTGTGCCGCCGCGGCCGCTGTCGCAACACCGCCGGCTCCTACACGTGCCTGTGCGAGCCCGGCTTCGCCCCCACAGCGGCCGGCTACTGCGCCGACGTGGACGAGTGCGCCGCCGACTCCATGTGCAAGGGCGGGCGCTGCGTGAACAGCGAGGGCTCGTTCCAATGCGTGTGCGAAGCGGGGTACCGGCCCACCGCCACCCGCGGCGCCTGCGTGGACGTGGACGAGTGCGAGGAGCAGCGCGTCTGCCGCAACGGCCGCTGCCACAACACGCCCGGCTCCTTCCGCTGCGAGTGCCTGCCCGGCTTCACGCTCAGCAACGACGGCAGGACGTGTCTAGATGAGGTTCAAGACTTATGTTACGAGAAATACGAAGAGGGCCACTGCACGGGGCCGGGACAGAACGCGGTCACCAGGTCGCAGTGCTGCTGCAGCGCCACTAAAGGTCTAAAGCTAGGCTGGGGCGTGTCGTGCCAGGCGTGCCCGGCGCAGGGCAGCGAGCTGTACGCGGTGCTGTGCCCCGAGGGACCCAGCAAGGACAACGGCGGCGCCGACATCAACGAGTGCACCGCGGTGCCCGGCGCCTGCCCGCACGGCTCCTGCGAGAACCTGGAGCCCGGCTACCGATGCATCTGCGACCCCGGCTACCATCCAGACGCGGACGGACTGTGCCGGGACATCGACGAGTGCGATATGCACCAATCCTACTGCGCGGGCGGCCAGTGCCGCAACACGGCGGGCAGCTTCACGTGCGTGTGTCCCGAGGGCACGCGCCACGAGCCCGAGCTGCAGCTGTGTCGGGACATCGACGAGTGCGAGGAGCTGGCGAGCCCGTGCGACAACGGACGCTGCATCAACACGCACGGCGGCTTCGAGTGCGAGTGCGAGCTGGGCTACGTGCTGGACGCCACCGCGCAGCGTTGTCTGGACAACCGGCGCGGCTCGTGCTGGCGCCGCGTGGTGGACGGACAGTGCGAGGCCGCCGCGCCAGAGCTACTCCTGCGCCAGGAGTGCTGCTGCAGCGTGGGGCTGGCCTGGGGCTCGCCCTGTGCGCCGTGCGATCCCGACCACTGCCCCTGCCCCAAGGGATTCGCCAAGCTGGACGGCGCCACGTGCCGCGACGTGGACGAGTGCGCGCTGAGTGCGGAGCTGTGCGCGGGCGGCACGTGCGTCAACACGGACGGCTCCTACCGCTGCGACTGCCCGCCCGGACTCACGCTGGACTCCACCGGTAACCGCTGCGTGGACCGGCGGCGCGAGTCCTGCTACACGGAGATGACGGGCGGGCGCTGCTCGGGCCCTCTACCCACCGACGTGCTGCGGGCCGTGTGCTGCTGTTCAGCCCTGGGCAAGGCCTGGGGGAACCAGCGCTGCGAGCCCTGCCCTAAGAAAGGAACGGATGCCTATCAAAACCTGTGCATAGCAGTGGGCACTCTGCTACCGCCGAACCAGTGGGACAGACCGATCGACGGCAACATGACCGACATCTGGTCACAGGTCGGAGGAGACGAGAACGGGGACGGGTATGGGAAACAGTGGGGTGAGAATGGTAGCGACGGAGGTTGGGATACCTGGGGGAACGGAACCGTTGGAGGAAACGGACCGGGACTGATCCCCGGACAGATGGAAGTTAACGAGTGTGCAGCTTTCCCTGGACTCTGCGGCCATGGACGCTGTAGAAACCTGCTTGGCGGGTTCACTTGTGATTGTTTCCCTGGATACGAAAAGGATTCAAAGAACCACACGTGCGTGGACGTGAACGAGTGTGAGATCGTGGACGGCGTGTGTGGTGACGGCGACTGTCACAACACTGAAGGCAGCTTCACCTGCCACTGCCGTCCTGGGTACAAGGCGGACGACTTCTCCAAGATATGTGTCGACATAGACGAGTGCGCGGACAACGACGCGTTGTGTCGCGGCGGCCGCTGCGTCAACACGGCGGGCTCGTTCCGCTGCGAGTGCGGCCACGGCATGGAGCTGGCGCCCGACAGACTCTCCTGCAAGGACATCGACGAGTGCTCCATCACCTCAGGTATCTGCAGCAACGGCGCGTGCGAGAACCAGATGGGCACGTACCAGTGCGTGTGCGACGAGGGCTACGCGCAGTCCACCGTGAAGTCGCACTGCGACGACATCGACGAGTGCGCCGAGGACGCCGCGCGCTGCCAGCACTCCTGCGTCAACACGCCCGGCTCCTACCACTGCACCTGCCGCGAGGGCTGGCACCTGCGCGCGGACGGGCGCTCGTGCCGCGACATCGACGAGTGCGCGGGCGGCGCGCGGCCCTGCGGCGGCGGACAGTGCCGCAACACGGTCGGCTCCTACACCTGCACCTGCACGGACGGGCTGGTGCCGTCGGCCGCCGGCGCCAAGCCCACCTGCCAGGACATCGACGAGTGCGCAGACATACCGGAGCTGTGCGGTGCCGGGTCGTGCCACAACACGATCGGCTCGTTCGTGTGCCAGTGTCCGGATGGGTACAGCGTGAAGCCGGAGCAGGGACCCGCCTGCACCGACGACGACGAGTGCGAGCTGGGCACCTGCGACTGCCACCCCGCCGCCGACTGCATCAACCTACCGGGCTCGTTCCAGTGCCGGTGTCGGGACGGGTGGCGCGGGGACGGCGCCACGTGCGAGGACGTCGACGAGTGCCTCACCAACAACGGCGGCTGCCACCCGCGGGCCACCTGCGCCAACACGGACGGCTCCTTCCGCTGCCTCTGCGACACCGGCTACAAGGGAGACGGGTACTCGTGCGTGGACATCGATGAGTGCGCCAACGACCCCACGCTGTGCGAGAACGGGCACTGCAGCAACACGCCGGGCGGCTACGAGTGCGACTGCGACGTGGGCTTCACGCGCGCCGCCGACGGACGCTCGTGTCTGGACATGGACGAGTGCGCCACGTTCGACAACGTTTGCGTATTCGGGCGTTGCGTGAACACGTACGGCATGTTCAAATGTATCTGCGACAAGGGGTACCAATCGGACAGCGTCGACGATCTGATGCCCGGCTTCAACTGCACGGACGTGGACGAGTGCAAGTCGCCGCAGTCGTGCCAGTACGGGCAGTGCATCAACACGCAGGGCTCCTACACGTGCCGCTGTCCGCCCAACTACGAGCTGGTCTCCGACGGCACGGCCTGCTACGATTCCCGTAAGGCCCGTTGCTACGGTAAAGTGGATCTGCGCTCCGGGACGGAGACATGCCGGGACAGCGATGAGCTCTCCGAGGACGGAACCATGGCCGCCTGTTGCTGTTCCGTCGGAGCGGCGTGGGGCAACTACTGCGACCCGTGTCCGGAGCCGGGCTCTGAGGCCTACCGCCAGCTGTGTCCTGGGGGACCGGGGTACCAGCCGGTACTAGAACCGCCGTCGTACGTGGTGACGCTGGCCGACATCGACGAGTGCGCCGCGCACGAGGGCCTGTGCGCGCACGGCACGTGCACCAACACGTTCGGCTCGTTCGTGTGCACGTGCGGCGCGGGCTGGAGTCTGGCGCCCGACGAGCTGAGCTGCGAGGACGACGACGAGTGCGAGCGCCCCGACGTGTGCGGGCCCGGGGTCTGCCGCAACCTGCCCGGCTCCTACGTGTGTCTGTGCCCAGAGGGATACGTCGCCATGCCCAACGGAAAGGAATGCGTGGACGTGAGACAACGACAATGCTACATGGAGTGGAACGAGGAGGACCAGAGCTGTGCGGGGGCGGTGGGAGTCCCGCAGACCAAGTACTTGTGCTGCTGCTCTGTGGGGCACGCCTGGGGAGCGCCGTGCGAGGCGTGTCCCCCGCGGGGGTCCCCGGCACACACGGCCCTGTGCGGCGAGAAGCCAGGGGAGTACATCAACCCTGTCACCAATGAGACGAAGCCCATCGACGAGTGCGACATCATGCCGCAGCTCTGCAAGCCTGGTACCTGCCACGACACGCCCACTGGGTTCCAGTGCGGATGTGATCATGGGTACGAGCACGACAACACATCGCACCTGTGCCGGGACGTGGACGAGTGCTCGTGGGGCCGGCCGCCCTGCCGCGGCATGGCGCAGTGCGTGAACCTGCCGGGCGCCTTCGAGTGCCGCTGCCCGCCCGGGTACCGCCTCACGCCCTCTCTGGACGAGTGCGAGGACGTGGACGAGTGTGGCGACCAGCGCATTTGCGACCACGGGGACTGCAGGAACACTATCGGCTCTTACAGATGCGAATGTAAGCCCGGGTACACTCTCCGGGAGAACGTGTGTCGCGACGTGGACGAGTGCTCGCGGCCGCGGCCCGTCTGCAGGAACGGCACGTGCGAGAACCTGCCGGGCGCGTACCTGTGCCACTGCGACGACGGCTTTAAGGCGGGACCCAACAACGACTGTGTCGATGTGAACGAGTGTCGTGAAGGCGGCATGGTCTGCCGCAACGGTCGCTGCCGGAACACGGTGGGCTCGTTCCGCTGCGAGTGTGCGCCCGGGTACACGCTCACGGCCGACGGACGCAACTGCCGCGATGTAGACGAGTGCGACGAACTGCCGCACCCCTGCGGGAGGGACGGGAATCCCTCTTGCACTAACACTAATGGAGGATACGAATGTTCATGTGGCGCGGGCTGGAAGTTAGTAGGGCGGCGATGTGTGGACCGCGACGAATGTAAGGAGCTGCCCTACGTGTGCGCCGGAGGAGAGTGCCGAAACTTTAATGGAGGATACGTTTGCGAGTGTCCGGCGGGCTGGCGGTTCGACAAGACGGCGGCCGTGTGCGTGGACGAGCGCAAGGAGCTCTGCTACGACGAGTGGGAGGCCGGCCGCTGCCACCGCGCGCGCCCCCTGCAGCTCGCGAGGCCGGAGTGCTGCTGCTCTGAGGGTGCCGCTTGGGGCCGTTACTGTGAGCGTTGTCCGGCGCCGGACTCCGCTGAGTTCATGAGGATCTGTCAAGGTGGAATGGGAAGACCCAATCTTACACAGGATCTAGACGAGTGTCGCGTCCGCCCCGACGTGTGCGTGGGCGGGCGGTGCGTCAATACGGACGGGTCGTTCCGCTGCGAGTGCCCGGACGGCTACGTGCTCGCCCCGGACGGGCTGTCCTGCGTGGACGCGGACGAGTGCGCCCTGGACCCCAGGATCTGCGGCAACGGGACCTGCTCCAACACGCGCGGAGGCTACGAGTGTCAGTGCAGCCCTGGCTTTACGCAGGGGCCGGATCAGACGTGCGTGGACGTGGACGAGTGCGCGGAGGGGCGCGCGTCGTGCACGTTCCGGTGCCACAACACGGCCGGCTCGTTCCGCTGCACGTGCCCGTACGGGTACGCCGTGGCGGCGGACGGCGTGCACTGCCGCGACATCGACGAGTGCGTGCAGGAGCCGCGCGTGTGTCCGCACGCCTGCGAGAACGTCGTAGGCTCCTACATCTGCAAGTGTCCGGAAGGATATCGTCGTACCTCGGCCCCGCAGGACTCTGAGAATGCCTGCGAGGATATCAACGAGTGCGAAGAACAAGAAGACCTATGTTCGGGAGGAGTCTGCATCAATACAGACGGGAGCTTCCTCTGTGACTGCGACGCCGGGTTCGAACCCAGCGAAGATGGAACCGACTGTATTGACCGTCGCACGGGCACGTGCTACCGCTCGCTGGTGTCGGGGCGCTGCGAGCCGGAACCCTGGCCGCGCGCCGCCCCCGCCACGTCCGCCGCGCCCGCCGCCTCCCCCGGCCCCCCCGCGCCCGGACACGTCACCAAGGCTCAGTGCTGCTGTACCTTGGGAGCGGCGTGGGGATCAGAATGCGAACTGTGTCCTGCACCCGGCTCCCCGGCGAGACTCGATCTGTGTACCGCGGTGAACTTGACCCCCGGGGGACAAGGAGGCCACGGAGGAGGCGGGGGCTCCGGAGGGTCGCATGGGGGCTCGGGCGGGTCGTTAGATGTGTTCGCTGATGTTGACGAGTGTGCCGCAATCCCAGGGCTGTGTGCACCCGGACGGTGTGTCAACACTATTGGAAGTTTCCGTTGCGTGTGCGGGCCCGGGTACCGTCCGGCGGGCGAGGTGTGCGCGGACGTGGACGAGTGCGCGGTCCGGCCGCCGCCGTGCGACCAGCTGTGCCACAACACGGACGGCTCGTACGACTGTCTCTGTCGCACCGGCTACGAGCTGGACGAGGACGGCGCCAACTGCCGCGACGTGGACGAGTGCGAGCGAGACACACATACCTGCCAGCAGATCTGCTCCAACACCGAGGGCTCCTATGAGTGCTCGTGCGAGGACGGGTATGAGAAGAGGGGTGATGCTTGTGTCGATATAAACGAGTGTCACGAGGAGGGCGTGTGTCCGTCCCCGGGGCGGTGCGTGAACCTGCTGGGCTCGTTCCGCTGCGTGTGTCCGCGCGGCTACCGGCTGGACGCGGAGGGCGCGCGGTGCGTGGACCGCGACGAGTGCGCGCAGACGGGCCGCTGCCAGGCGCCCTGCCGGAACTACGCCGGCGGCTACCGCTGCGACTGTCCCGTCGGCACGGTGCGGTCGCCGAGCGGGGCCTGCGTGCCGGCGGACCCGTGTTCGAACGCGGAGTGTGGCGCGTCCCCTTGCTTCCCGGTGGGAGGGGTCTACCGCTGCGGCTGTCCCCCGGGGTACGGCTGGGATGCCTCTCATGCTGTCTGCCTACAGCTGGGCGGCGGCTGCGCCACAGCGAACTGTCTGTTCGGGTGCCGGTCGCTCGGGTCCTCGTTCGAGTGCGGCTGTCCGGGCGGGTACGCGCTGGTGGCCGGCCACTGTCTGGCGGGCGCGGACGGCTCGCTGTCCCCGGACGACATCGGGGACGCGCCCGTGTTCCCGCTGCACGACCAGTACAAGCTGGGCGGCCAGCAGGACCTCATCTCCACCGAAGGCTGCTTCAGTTGTAAGGTGAACGGCCGACACCGGAGAACACCGGACGAAGCCGTCATCTACGCGAACGGAACGACGCTCATGAGGAAACGGAAACGGCGCAGTCGAAGTCGCAGGTCAGTCTTGGAACCGGAAGCGGAGCTGGTGGTGGTCCGGTCGCGGCCGGAGCGCACGTGGGCCCGCGCCCCCCTGCTGCGCCTCACCCCCGCCGCGCCCCGCCCCCGCCCCCACTACCGCATCGCGTACGGCGACCCGGACCGCCTGTTCACGCTGTCCAAGCGGGACGGCGCCTGGGCGCTGCGGCTCCGCAAGCAGCTGCCCCGGGACGCCGAGCTGCGCGCGCAGCTGGAGATCGAGGCCCGCTTCGTGTCCCCTCGCCGCATGAGGCGTGACGTCACGTCCTCGCAGCCGCCGGTCAGACTGTACGTGTCCGTTGACATTTCACCAGACCACAAGCACTGA
Protein
MSYWRLLVAAALLASVAGDPRTPRGTTRPRHWQRGQRMLDDRAPSQFVGSHVCRSRNSTHCCPGWTSRPNSLLCIVPLCRPDCGSPGACIAPNICRCPGGVEAPSCGNGGLYPYPVRTQGGCRRICMNGGTCTNGTCACAPGWSGEFCTEPICKEPCLHGGRCIAPDRCVCFHGLSGPRCEIDRRTGPCYTETRGALCTGALEGVVCTRQLCCATVGRAWGHPCERCSDLDCPVGHLRNLATKECQDIDECAAVPGLCSGGRCINSIGSFSCECPPGQRRHPLSNNCEDIDECEDPDICPNGKCVNTEGDYYCLCNPHFIPSPDKKFCIDGRVGSCYAYLSETGECADRLPVALSNRDCCCGYNMGRAWGDLCTPCPPRGATEWTHLCGGGAIPPNWRRNVTGGGGGGGIGGGGQGGEGGGSSGGGEDGDGGGGTFAPPSIAKINECMLRNGICGLGNCIDKDVGYTCECWAGAEVAEIDGNRVCIDIDECALEYCKGGTCINRPGDFECRCPPGFDPVENGLRCSDKNECEMTNGGMCTNGICKNIDGGFECICKPGYESGSEGALCRDVDECRDNPRVCRRGRCRNTAGSYTCLCEPGFAPTAAGYCADVDECAADSMCKGGRCVNSEGSFQCVCEAGYRPTATRGACVDVDECEEQRVCRNGRCHNTPGSFRCECLPGFTLSNDGRTCLDEVQDLCYEKYEEGHCTGPGQNAVTRSQCCCSATKGLKLGWGVSCQACPAQGSELYAVLCPEGPSKDNGGADINECTAVPGACPHGSCENLEPGYRCICDPGYHPDADGLCRDIDECDMHQSYCAGGQCRNTAGSFTCVCPEGTRHEPELQLCRDIDECEELASPCDNGRCINTHGGFECECELGYVLDATAQRCLDNRRGSCWRRVVDGQCEAAAPELLLRQECCCSVGLAWGSPCAPCDPDHCPCPKGFAKLDGATCRDVDECALSAELCAGGTCVNTDGSYRCDCPPGLTLDSTGNRCVDRRRESCYTEMTGGRCSGPLPTDVLRAVCCCSALGKAWGNQRCEPCPKKGTDAYQNLCIAVGTLLPPNQWDRPIDGNMTDIWSQVGGDENGDGYGKQWGENGSDGGWDTWGNGTVGGNGPGLIPGQMEVNECAAFPGLCGHGRCRNLLGGFTCDCFPGYEKDSKNHTCVDVNECEIVDGVCGDGDCHNTEGSFTCHCRPGYKADDFSKICVDIDECADNDALCRGGRCVNTAGSFRCECGHGMELAPDRLSCKDIDECSITSGICSNGACENQMGTYQCVCDEGYAQSTVKSHCDDIDECAEDAARCQHSCVNTPGSYHCTCREGWHLRADGRSCRDIDECAGGARPCGGGQCRNTVGSYTCTCTDGLVPSAAGAKPTCQDIDECADIPELCGAGSCHNTIGSFVCQCPDGYSVKPEQGPACTDDDECELGTCDCHPAADCINLPGSFQCRCRDGWRGDGATCEDVDECLTNNGGCHPRATCANTDGSFRCLCDTGYKGDGYSCVDIDECANDPTLCENGHCSNTPGGYECDCDVGFTRAADGRSCLDMDECATFDNVCVFGRCVNTYGMFKCICDKGYQSDSVDDLMPGFNCTDVDECKSPQSCQYGQCINTQGSYTCRCPPNYELVSDGTACYDSRKARCYGKVDLRSGTETCRDSDELSEDGTMAACCCSVGAAWGNYCDPCPEPGSEAYRQLCPGGPGYQPVLEPPSYVVTLADIDECAAHEGLCAHGTCTNTFGSFVCTCGAGWSLAPDELSCEDDDECERPDVCGPGVCRNLPGSYVCLCPEGYVAMPNGKECVDVRQRQCYMEWNEEDQSCAGAVGVPQTKYLCCCSVGHAWGAPCEACPPRGSPAHTALCGEKPGEYINPVTNETKPIDECDIMPQLCKPGTCHDTPTGFQCGCDHGYEHDNTSHLCRDVDECSWGRPPCRGMAQCVNLPGAFECRCPPGYRLTPSLDECEDVDECGDQRICDHGDCRNTIGSYRCECKPGYTLRENVCRDVDECSRPRPVCRNGTCENLPGAYLCHCDDGFKAGPNNDCVDVNECREGGMVCRNGRCRNTVGSFRCECAPGYTLTADGRNCRDVDECDELPHPCGRDGNPSCTNTNGGYECSCGAGWKLVGRRCVDRDECKELPYVCAGGECRNFNGGYVCECPAGWRFDKTAAVCVDERKELCYDEWEAGRCHRARPLQLARPECCCSEGAAWGRYCERCPAPDSAEFMRICQGGMGRPNLTQDLDECRVRPDVCVGGRCVNTDGSFRCECPDGYVLAPDGLSCVDADECALDPRICGNGTCSNTRGGYECQCSPGFTQGPDQTCVDVDECAEGRASCTFRCHNTAGSFRCTCPYGYAVAADGVHCRDIDECVQEPRVCPHACENVVGSYICKCPEGYRRTSAPQDSENACEDINECEEQEDLCSGGVCINTDGSFLCDCDAGFEPSEDGTDCIDRRTGTCYRSLVSGRCEPEPWPRAAPATSAAPAASPGPPAPGHVTKAQCCCTLGAAWGSECELCPAPGSPARLDLCTAVNLTPGGQGGHGGGGGSGGSHGGSGGSLDVFADVDECAAIPGLCAPGRCVNTIGSFRCVCGPGYRPAGEVCADVDECAVRPPPCDQLCHNTDGSYDCLCRTGYELDEDGANCRDVDECERDTHTCQQICSNTEGSYECSCEDGYEKRGDACVDINECHEEGVCPSPGRCVNLLGSFRCVCPRGYRLDAEGARCVDRDECAQTGRCQAPCRNYAGGYRCDCPVGTVRSPSGACVPADPCSNAECGASPCFPVGGVYRCGCPPGYGWDASHAVCLQLGGGCATANCLFGCRSLGSSFECGCPGGYALVAGHCLAGADGSLSPDDIGDAPVFPLHDQYKLGGQQDLISTEGCFSCKVNGRHRRTPDEAVIYANGTTLMRKRKRRSRSRRSVLEPEAELVVVRSRPERTWARAPLLRLTPAAPRPRPHYRIAYGDPDRLFTLSKRDGAWALRLRKQLPRDAELRAQLEIEARFVSPRRMRRDVTSSQPPVRLYVSVDISPDHKH
Summary
Description
Fibrillin-2: Fibrillins are structural components of 10-12 nm extracellular calcium-binding microfibrils, which occur either in association with elastin or in elastin-free bundles. Fibrillin-2-containing microfibrils regulate the early process of elastic fiber assembly. Regulates osteoblast maturation by controlling TGF-beta bioavailability and calibrating TGF-beta and BMP levels, respectively.
Fibrillin-2: Fibrillins are structural components of 10-12 nm extracellular calcium-binding microfibrils, which occur either in association with elastin or in elastin-free bundles. Fibrillin-2-containing microfibrils regulate the early process of elastic fiber assembly. Regulates osteoblast maturation by controlling TGF-beta bioavailability and calibrating TGF-beta and BMP levels, respectively (PubMed:20855508).
Fibrillin-1: Structural component of the 10-12 nm diameter microfibrils of the extracellular matrix, which conveys both structural and regulatory properties to load-bearing connective tissues (PubMed:1860873, PubMed:15062093). Fibrillin-1-containing microfibrils provide long-term force bearing structural support. In tissues such as the lung, blood vessels and skin, microfibrils form the periphery of the elastic fiber, acting as a scaffold for the deposition of elastin. In addition, microfibrils can occur as elastin-independent networks in tissues such as the ciliary zonule, tendon, cornea and glomerulus where they provide tensile strength and have anchoring roles. Fibrillin-1 also plays a key role in tissue homeostasis through specific interactions with growth factors, such as the bone morphogenetic proteins (BMPs), growth and differentiation factors (GDFs) and latent transforming growth factor-beta-binding proteins (LTBPs), cell-surface integrins and other extracellular matrix protein and proteoglycan components (PubMed:27026396). Regulates osteoblast maturation by controlling TGF-beta bioavailability and calibrating TGF-beta and BMP levels, respectively (By similarity). Negatively regulates osteoclastogenesis by binding and sequestering an osteoclast differentiation and activation factor TNFSF11. This leads to disruption of TNFSF11-induced Ca(2+) signaling and impairment of TNFSF11-mediated nuclear translocation and activation of transcription factor NFATC1 which regulates genes important for osteoclast differentiation and function (PubMed:24039232). Mediates cell adhesion via its binding to cell surface receptors integrins ITGAV:ITGB3 and ITGA5:ITGB1 (PubMed:12807887, PubMed:17158881). Binds heparin and this interaction has an important role in the assembly of microfibrils (PubMed:11461921).
Asprosin: Hormone that targets the liver to increase plasma glucose levels. Secreted by white adipose tissue and circulates in the plasma. Acts in response to fasting and promotes blood glucose elevation by binding to the surface of hepatocytes. Promotes hepatocyte glucose release by activating the protein kinase A activity in the liver, resulting in rapid glucose release into the circulation.
Fibrillin-1: Structural component of the 10-12 nm diameter microfibrils of the extracellular matrix, which conveys both structural and regulatory properties to load-bearing connective tissues. Fibrillin-1-containing microfibrils provide long-term force bearing structural support. In tissues such as the lung, blood vessels and skin, microfibrils form the periphery of the elastic fiber, acting as a scaffold for the deposition of elastin. In addition, microfibrils can occur as elastin-independent networks in tissues such as the ciliary zonule, tendon, cornea and glomerulus where they provide tensile strength and have anchoring roles. Fibrillin-1 also plays a key role in tissue homeostasis through specific interactions with growth factors, such as the bone morphogenetic proteins (BMPs), growth and differentiation factors (GDFs) and latent transforming growth factor-beta-binding proteins (LTBPs), cell-surface integrins and other extracellular matrix protein and proteoglycan components (By similarity). Regulates osteoblast maturation by controlling TGF-beta bioavailability and calibrating TGF-beta and BMP levels, respectively (PubMed:20855508). Negatively regulates osteoclastogenesis by binding and sequestering an osteoclast differentiation and activation factor TNFSF11. This leads to disruption of TNFSF11-induced Ca(2+) signaling and impairment of TNFSF11-mediated nuclear translocation and activation of transcription factor NFATC1 which regulates genes important for osteoclast differentiation and function (PubMed:24039232). Mediates cell adhesion via its binding to cell surface receptors integrins ITGAV:ITGB3 and ITGA5:ITGB1. Binds heparin and this interaction plays an important role in the assembly of microfibrils (By similarity).
Fibrillin-1: Structural component of the 10-12 nm diameter microfibrils of the extracellular matrix, which conveys both structural and regulatory properties to load-bearing connective tissues. Fibrillin-1-containing microfibrils provide long-term force bearing structural support. In tissues such as the lung, blood vessels and skin, microfibrils form the periphery of the elastic fiber, acting as a scaffold for the deposition of elastin. In addition, microfibrils can occur as elastin-independent networks in tissues such as the ciliary zonule, tendon, cornea and glomerulus where they provide tensile strength and have anchoring roles. Fibrillin-1 also plays a key role in tissue homeostasis through specific interactions with growth factors, such as the bone morphogenetic proteins (BMPs), growth and differentiation factors (GDFs) and latent transforming growth factor-beta-binding proteins (LTBPs), cell-surface integrins and other extracellular matrix protein and proteoglycan components. Regulates osteoblast maturation by controlling TGF-beta bioavailability and calibrating TGF-beta and BMP levels, respectively. Negatively regulates osteoclastogenesis by binding and sequestering an osteoclast differentiation and activation factor TNFSF11. This leads to disruption of TNFSF11-induced Ca(2+) signaling and impairment of TNFSF11-mediated nuclear translocation and activation of transcription factor NFATC1 which regulates genes important for osteoclast differentiation and function. Mediates cell adhesion via its binding to cell surface receptors integrins ITGAV:ITGB3 and ITGA5:ITGB1. Binds heparin and this interaction plays an important role in the assembly of microfibrils.
Fibrillin-3: Fibrillins are structural components of 10-12 nm extracellular calcium-binding microfibrils, which occur either in association with elastin or in elastin-free bundles. Fibrillin-containing microfibrils provide long-term force bearing structural support.
May play an integral structural role in elastic-fiber architectural organization and/or assembly.
Key regulator of transforming growth factor beta (TGFB1, TGFB2 and TGFB3) that controls TGF-beta activation by maintaining it in a latent state during storage in extracellular space. Associates specifically via disulfide bonds with the Latency-associated peptide (LAP), which is the regulatory chain of TGF-beta, and regulates integrin-dependent activation of TGF-beta. Outcompeted by LRRC32/GARP for binding to LAP regulatory chain of TGF-beta.
Key regulator of transforming growth factor beta (TGFB1, TGFB2 and TGFB3) that controls TGF-beta activation by maintaining it in a latent state during storage in extracellular space. Associates specifically via disulfide bonds with the Latency-associated peptide (LAP), which is the regulatory chain of TGF-beta, and regulates integrin-dependent activation of TGF-beta.
Key regulator of transforming growth factor beta (TGFB1, TGFB2 and TGFB3) that controls TGF-beta activation by maintaining it in a latent state during storage in extracellular space (PubMed:7593177). Associates specifically via disulfide bonds with the Latency-associated peptide (LAP), which is the regulatory chain of TGF-beta, and regulates integrin-dependent activation of TGF-beta (By similarity). Outcompeted by LRRC32/GARP for binding to LAP regulatory chain of TGF-beta (By similarity).
Key regulator of transforming growth factor beta (TGFB1, TGFB2 and TGFB3) that controls TGF-beta activation by maintaining it in a latent state during storage in extracellular space (PubMed:2022183, PubMed:8617200, PubMed:8939931). Associates specifically via disulfide bonds with the Latency-associated peptide (LAP), which is the regulatory chain of TGF-beta, and regulates integrin-dependent activation of TGF-beta (PubMed:8617200, PubMed:8939931, PubMed:15184403). Outcompeted by LRRC32/GARP for binding to LAP regulatory chain of TGF-beta (PubMed:22278742).
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs. Involved in angiogenesis; negatively regulates endothelial cell proliferation and migration and angiogenic sprouting. Involved in the maturation of both CD4(+) and CD8(+) cells in the thymus. Important for follicular differentiation and possibly cell fate selection within the follicle. During cerebellar development, functions as a receptor for neuronal DNER and is involved in the differentiation of Bergmann glia. Represses neuronal and myogenic differentiation. May play an essential role in postimplantation development, probably in some aspect of cell specification and/or differentiation. May be involved in mesoderm development, somite formation and neurogenesis. Involved in determination of left/right symmetry by modulating the balance between motile and immotile (sensory) cilia at the left-right organiser (LRO) (By similarity).
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs. Involved in angiogenesis; negatively regulates endothelial cell proliferation and migration and angiogenic sprouting. Involved in the maturation of both CD4(+) and CD8(+) cells in the thymus. Important for follicular differentiation and possibly cell fate selection within the follicle. During cerebellar development, functions as a receptor for neuronal DNER and is involved in the differentiation of Bergmann glia. Represses neuronal and myogenic differentiation. May play an essential role in postimplantation development, probably in some aspect of cell specification and/or differentiation. May be involved in mesoderm development, somite formation and neurogenesis. May enhance HIF1A function by sequestering HIF1AN away from HIF1A. Required for the THBS4 function in regulating protective astrogenesis from the subventricular zone (SVZ) niche after injury. Involved in determination of left/right symmetry by modulating the balance between motile and immotile (sensory) cilia at the left-right organiser (LRO).
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs. Involved in angiogenesis; negatively regulates endothelial cell proliferation and migration and angiogenic sprouting. Involved in the maturation of both CD4(+) and CD8(+) cells in the thymus. Important for follicular differentiation and possibly cell fate selection within the follicle. During cerebellar development, functions as a receptor for neuronal DNER and is involved in the differentiation of Bergmann glia. Represses neuronal and myogenic differentiation. May play an essential role in postimplantation development, probably in some aspect of cell specification and/or differentiation. May be involved in mesoderm development, somite formation and neurogenesis. May enhance HIF1A function by sequestering HIF1AN away from HIF1A (By similarity). Required for the THBS4 function in regulating protective astrogenesis from the subventricular zone (SVZ) niche after injury. Involved in determination of left/right symmetry by modulating the balance between motile and immotile (sensory) cilia at the left-right organiser (LRO) (By similarity).
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs. Involved in angiogenesis; negatively regulates endothelial cell proliferation and migration and angiogenic sprouting. Involved in the maturation of both CD4(+) and CD8(+) cells in the thymus. Important for follicular differentiation and possibly cell fate selection within the follicle. During cerebellar development, functions as a receptor for neuronal DNER and is involved in the differentiation of Bergmann glia. Represses neuronal and myogenic differentiation. May play an essential role in postimplantation development, probably in some aspect of cell specification and/or differentiation. May be involved in mesoderm development, somite formation and neurogenesis (By similarity). Involved in determination of left/right symmetry by modulating the balance between motile and immotile (sensory) cilia at the left-right organiser (LRO).
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination (By similarity). Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus (By similarity). Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity). Involved in angiogenesis; negatively regulates endothelial cell proliferation and migration and angiogenic sprouting (By similarity). Involved in the maturation of both CD4(+) and CD8(+) cells in the thymus (By similarity). Important for follicular differentiation and possibly cell fate selection within the follicle (By similarity). During cerebellar development, functions as a receptor for neuronal DNER and is involved in the differentiation of Bergmann glia (PubMed:11182080). Represses neuronal and myogenic differentiation (By similarity). May play an essential role in postimplantation development, probably in some aspect of cell specification and/or differentiation (By similarity). May be involved in mesoderm development, somite formation and neurogenesis (By similarity). May enhance HIF1A function by sequestering HIF1AN away from HIF1A (By similarity). Required for the THBS4 function in regulating protective astrogenesis from the subventricular zone (SVZ) niche after injury (By similarity). Involved in determination of left/right symmetry by modulating the balance between motile and immotile (sensory) cilia at the left-right organiser (LRO) (By similarity).
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus (PubMed:21378985, PubMed:21378989). Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity). Involved in bone remodeling and homeostasis. In collaboration with RELA/p65 enhances NFATc1 promoter activity and positively regulates RANKL-induced osteoclast differentiation (PubMed:29149593). Positively regulates self-renewal of liver cancer cells (PubMed:25985737).
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination (PubMed:10393120). Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus (PubMed:10393120, PubMed:18710934). Affects the implementation of differentiation, proliferation and apoptotic programs (PubMed:10393120, PubMed:18710934). May play an essential role in postimplantation development, probably in some aspect of cell specification and/or differentiation (By similarity). In collaboration with RELA/p65 enhances NFATc1 promoter activity and positively regulates RANKL-induced osteoclast differentiation (PubMed:18710934). Positively regulates self-renewal of liver cancer cells (By similarity).
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity). Involved in bone remodeling and homeostasis. In collaboration with RELA/p65 enhances NFATc1 promoter activity and positively regulates RANKL-induced osteoclast differentiation. Positively regulates self-renewal of liver cancer cells (By similarity).
Functions as a receptor for membrane-bound ligands Jagged1, Jagged2 and Delta1 to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity). May play a role during CNS development.
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs. Involved in angiogenesis; negatively regulates endothelial cell proliferation and migration and angiogenic sprouting. Involved in the maturation of both CD4(+) and CD8(+) cells in the thymus. Important for follicular differentiation and possibly cell fate selection within the follicle. During cerebellar development, functions as a receptor for neuronal DNER and is involved in the differentiation of Bergmann glia. Represses neuronal and myogenic differentiation. May play an essential role in postimplantation development, probably in some aspect of cell specification and/or differentiation. May be involved in mesoderm development, somite formation and neurogenesis (By similarity). Involved in determination of left/right symmetry by modulating the balance between motile and immotile (sensory) cilia at the left-right organiser (LRO) (By similarity).
Essential signaling protein which has a major role in many developmental processes (PubMed:3935325). Functions as a receptor for membrane-bound ligands Delta and Serrate to regulate cell-fate determination (PubMed:10935637, PubMed:15620650, PubMed:12909620, PubMed:18243100). Upon ligand activation, and releasing from the cell membrane, the Notch intracellular domain (NICD) forms a transcriptional activator complex with Su(H) (Suppressor of hairless) and activates genes of the E(spl) complex (PubMed:7671825). Regulates oogenesis, the differentiation of the ectoderm and the development of the central and peripheral nervous system, eye, wing disk, muscles and segmental appendages such as antennae and legs, through lateral inhibition or induction (PubMed:11719214, PubMed:12369105, PubMed:3935325). Regulates neuroblast self-renewal, identity and proliferation through the regulation of bHLH-O proteins; in larval brains, involved in the maintenance of type II neuroblast self-renewal and identity by suppressing erm expression together with pnt; might also regulate dpn expression through the activation of the transcriptional regulator Su(H) (PubMed:27151950, PubMed:28899667, PubMed:20152183, PubMed:18342578, PubMed:23056424, PubMed:21262215).
Functions as a receptor for membrane-bound ligands Jagged1, Jagged2 and Delta1 to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity). Acts instructively to control the cell fate determination of CNS multipotent progenitor cells, resulting in astroglial induction and neuron/oligodendrocyte suppression.
Functions as a receptor for membrane-bound ligands Jagged1, Jagged2 and Delta1 to regulate cell-fate determination (PubMed:15350543). Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity).
Required to maintain the integrity of photoreceptor cells (PubMed:27737822, PubMed:28378834, PubMed:30052645). Specifically required for normal morphology of the photoreceptor ciliary pocket, and might thus facilitate protein trafficking between the photoreceptor inner and outer segments via the transition zone (PubMed:27737822).
Functions as a receptor for membrane-bound ligands Jagged1, Jagged2 and Delta1 to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs. May regulate branching morphogenesis in the developing vascular system (By similarity).
Functions as a receptor for membrane-bound ligands Jagged1, Jagged2 and Delta1 to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity). May regulate branching morphogenesis in the developing vascular system.
Cell surface receptor that binds to the chondroitin sulfate moiety of glycosaminoglycan chains and promotes cell attachment. Promotes granulocyte chemotaxis, degranulation and adhesion. In macrophages, promotes the release of inflammatory cytokines, including IL8 and TNF. Signals probably through G-proteins.
Plays a central role in cell polarity establishment (PubMed:2344615, PubMed:12900452, PubMed:10102271, PubMed:11740560). Participates in the assembly, positioning and maintenance of adherens junctions via its interaction with the SAC complex (PubMed:11740560, PubMed:12900452, PubMed:10102271, PubMed:11076972). Controls the coalescence of the spots of zonula adherens (ZA) into a adhesive ring around the cells (PubMed:11740560). It may act as a signal (PubMed:2344615). Involved in morphogenesis of the photoreceptor rhabdomere, for positioning and growth of rhabdomere and AJ during the crucial period of photoreceptor extension along the proximodistal axis of the retina (PubMed:12900452). Component of the crb-galla-Xpd (CGX) complex which is essential for proper mitotic chromosome segregation in early embryos (PubMed:25065591). The CGX complex is also required for cell proliferation in developing wing disks (PubMed:25065591). In the CGX complex, acts with galla-1 or galla-2 to recruit Xpd and thus form the functional complex. Together with apn, plays a key role in trachea development at larval stages (PubMed:30645584).
Ligand for multiple Notch receptors and involved in the mediation of Notch signaling. May be involved in cell-fate decisions during hematopoiesis. Seems to be involved in early and late stages of mammalian cardiovascular development. Inhibits myoblast differentiation (By similarity). May regulate fibroblast growth factor-induced angiogenesis.
Functional ligand of integrin alpha-8/beta-1 in kidney development. Regulates the expression of GDNF with integrin alpha-8/beta-1 which is essential for kidney development. May also play a role in the development and function of various tissues, regulating cell adhesion, spreading and survival through the binding of several integrins (By similarity).
Ligand for multiple Notch receptors and involved in the mediation of Notch signaling (PubMed:18660822, PubMed:20437614). May be involved in cell-fate decisions during hematopoiesis (PubMed:9462510). Seems to be involved in early and late stages of mammalian cardiovascular development. Inhibits myoblast differentiation (By similarity). Enhances fibroblast growth factor-induced angiogenesis (in vitro).
Functional ligand of integrin alpha-8/beta-1 in kidney development. Regulates the expression of GDNF with integrin alpha-8/beta-1 which is essential for kidney development. May also play a role in the development and function of various tissues, regulating cell adhesion, spreading and survival through the binding of several integrins.
Fibrillin-2: Fibrillins are structural components of 10-12 nm extracellular calcium-binding microfibrils, which occur either in association with elastin or in elastin-free bundles. Fibrillin-2-containing microfibrils regulate the early process of elastic fiber assembly. Regulates osteoblast maturation by controlling TGF-beta bioavailability and calibrating TGF-beta and BMP levels, respectively (PubMed:20855508).
Fibrillin-1: Structural component of the 10-12 nm diameter microfibrils of the extracellular matrix, which conveys both structural and regulatory properties to load-bearing connective tissues (PubMed:1860873, PubMed:15062093). Fibrillin-1-containing microfibrils provide long-term force bearing structural support. In tissues such as the lung, blood vessels and skin, microfibrils form the periphery of the elastic fiber, acting as a scaffold for the deposition of elastin. In addition, microfibrils can occur as elastin-independent networks in tissues such as the ciliary zonule, tendon, cornea and glomerulus where they provide tensile strength and have anchoring roles. Fibrillin-1 also plays a key role in tissue homeostasis through specific interactions with growth factors, such as the bone morphogenetic proteins (BMPs), growth and differentiation factors (GDFs) and latent transforming growth factor-beta-binding proteins (LTBPs), cell-surface integrins and other extracellular matrix protein and proteoglycan components (PubMed:27026396). Regulates osteoblast maturation by controlling TGF-beta bioavailability and calibrating TGF-beta and BMP levels, respectively (By similarity). Negatively regulates osteoclastogenesis by binding and sequestering an osteoclast differentiation and activation factor TNFSF11. This leads to disruption of TNFSF11-induced Ca(2+) signaling and impairment of TNFSF11-mediated nuclear translocation and activation of transcription factor NFATC1 which regulates genes important for osteoclast differentiation and function (PubMed:24039232). Mediates cell adhesion via its binding to cell surface receptors integrins ITGAV:ITGB3 and ITGA5:ITGB1 (PubMed:12807887, PubMed:17158881). Binds heparin and this interaction has an important role in the assembly of microfibrils (PubMed:11461921).
Asprosin: Hormone that targets the liver to increase plasma glucose levels. Secreted by white adipose tissue and circulates in the plasma. Acts in response to fasting and promotes blood glucose elevation by binding to the surface of hepatocytes. Promotes hepatocyte glucose release by activating the protein kinase A activity in the liver, resulting in rapid glucose release into the circulation.
Fibrillin-1: Structural component of the 10-12 nm diameter microfibrils of the extracellular matrix, which conveys both structural and regulatory properties to load-bearing connective tissues. Fibrillin-1-containing microfibrils provide long-term force bearing structural support. In tissues such as the lung, blood vessels and skin, microfibrils form the periphery of the elastic fiber, acting as a scaffold for the deposition of elastin. In addition, microfibrils can occur as elastin-independent networks in tissues such as the ciliary zonule, tendon, cornea and glomerulus where they provide tensile strength and have anchoring roles. Fibrillin-1 also plays a key role in tissue homeostasis through specific interactions with growth factors, such as the bone morphogenetic proteins (BMPs), growth and differentiation factors (GDFs) and latent transforming growth factor-beta-binding proteins (LTBPs), cell-surface integrins and other extracellular matrix protein and proteoglycan components (By similarity). Regulates osteoblast maturation by controlling TGF-beta bioavailability and calibrating TGF-beta and BMP levels, respectively (PubMed:20855508). Negatively regulates osteoclastogenesis by binding and sequestering an osteoclast differentiation and activation factor TNFSF11. This leads to disruption of TNFSF11-induced Ca(2+) signaling and impairment of TNFSF11-mediated nuclear translocation and activation of transcription factor NFATC1 which regulates genes important for osteoclast differentiation and function (PubMed:24039232). Mediates cell adhesion via its binding to cell surface receptors integrins ITGAV:ITGB3 and ITGA5:ITGB1. Binds heparin and this interaction plays an important role in the assembly of microfibrils (By similarity).
Fibrillin-1: Structural component of the 10-12 nm diameter microfibrils of the extracellular matrix, which conveys both structural and regulatory properties to load-bearing connective tissues. Fibrillin-1-containing microfibrils provide long-term force bearing structural support. In tissues such as the lung, blood vessels and skin, microfibrils form the periphery of the elastic fiber, acting as a scaffold for the deposition of elastin. In addition, microfibrils can occur as elastin-independent networks in tissues such as the ciliary zonule, tendon, cornea and glomerulus where they provide tensile strength and have anchoring roles. Fibrillin-1 also plays a key role in tissue homeostasis through specific interactions with growth factors, such as the bone morphogenetic proteins (BMPs), growth and differentiation factors (GDFs) and latent transforming growth factor-beta-binding proteins (LTBPs), cell-surface integrins and other extracellular matrix protein and proteoglycan components. Regulates osteoblast maturation by controlling TGF-beta bioavailability and calibrating TGF-beta and BMP levels, respectively. Negatively regulates osteoclastogenesis by binding and sequestering an osteoclast differentiation and activation factor TNFSF11. This leads to disruption of TNFSF11-induced Ca(2+) signaling and impairment of TNFSF11-mediated nuclear translocation and activation of transcription factor NFATC1 which regulates genes important for osteoclast differentiation and function. Mediates cell adhesion via its binding to cell surface receptors integrins ITGAV:ITGB3 and ITGA5:ITGB1. Binds heparin and this interaction plays an important role in the assembly of microfibrils.
Fibrillin-3: Fibrillins are structural components of 10-12 nm extracellular calcium-binding microfibrils, which occur either in association with elastin or in elastin-free bundles. Fibrillin-containing microfibrils provide long-term force bearing structural support.
May play an integral structural role in elastic-fiber architectural organization and/or assembly.
Key regulator of transforming growth factor beta (TGFB1, TGFB2 and TGFB3) that controls TGF-beta activation by maintaining it in a latent state during storage in extracellular space. Associates specifically via disulfide bonds with the Latency-associated peptide (LAP), which is the regulatory chain of TGF-beta, and regulates integrin-dependent activation of TGF-beta. Outcompeted by LRRC32/GARP for binding to LAP regulatory chain of TGF-beta.
Key regulator of transforming growth factor beta (TGFB1, TGFB2 and TGFB3) that controls TGF-beta activation by maintaining it in a latent state during storage in extracellular space. Associates specifically via disulfide bonds with the Latency-associated peptide (LAP), which is the regulatory chain of TGF-beta, and regulates integrin-dependent activation of TGF-beta.
Key regulator of transforming growth factor beta (TGFB1, TGFB2 and TGFB3) that controls TGF-beta activation by maintaining it in a latent state during storage in extracellular space (PubMed:7593177). Associates specifically via disulfide bonds with the Latency-associated peptide (LAP), which is the regulatory chain of TGF-beta, and regulates integrin-dependent activation of TGF-beta (By similarity). Outcompeted by LRRC32/GARP for binding to LAP regulatory chain of TGF-beta (By similarity).
Key regulator of transforming growth factor beta (TGFB1, TGFB2 and TGFB3) that controls TGF-beta activation by maintaining it in a latent state during storage in extracellular space (PubMed:2022183, PubMed:8617200, PubMed:8939931). Associates specifically via disulfide bonds with the Latency-associated peptide (LAP), which is the regulatory chain of TGF-beta, and regulates integrin-dependent activation of TGF-beta (PubMed:8617200, PubMed:8939931, PubMed:15184403). Outcompeted by LRRC32/GARP for binding to LAP regulatory chain of TGF-beta (PubMed:22278742).
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs. Involved in angiogenesis; negatively regulates endothelial cell proliferation and migration and angiogenic sprouting. Involved in the maturation of both CD4(+) and CD8(+) cells in the thymus. Important for follicular differentiation and possibly cell fate selection within the follicle. During cerebellar development, functions as a receptor for neuronal DNER and is involved in the differentiation of Bergmann glia. Represses neuronal and myogenic differentiation. May play an essential role in postimplantation development, probably in some aspect of cell specification and/or differentiation. May be involved in mesoderm development, somite formation and neurogenesis. Involved in determination of left/right symmetry by modulating the balance between motile and immotile (sensory) cilia at the left-right organiser (LRO) (By similarity).
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs. Involved in angiogenesis; negatively regulates endothelial cell proliferation and migration and angiogenic sprouting. Involved in the maturation of both CD4(+) and CD8(+) cells in the thymus. Important for follicular differentiation and possibly cell fate selection within the follicle. During cerebellar development, functions as a receptor for neuronal DNER and is involved in the differentiation of Bergmann glia. Represses neuronal and myogenic differentiation. May play an essential role in postimplantation development, probably in some aspect of cell specification and/or differentiation. May be involved in mesoderm development, somite formation and neurogenesis. May enhance HIF1A function by sequestering HIF1AN away from HIF1A. Required for the THBS4 function in regulating protective astrogenesis from the subventricular zone (SVZ) niche after injury. Involved in determination of left/right symmetry by modulating the balance between motile and immotile (sensory) cilia at the left-right organiser (LRO).
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs. Involved in angiogenesis; negatively regulates endothelial cell proliferation and migration and angiogenic sprouting. Involved in the maturation of both CD4(+) and CD8(+) cells in the thymus. Important for follicular differentiation and possibly cell fate selection within the follicle. During cerebellar development, functions as a receptor for neuronal DNER and is involved in the differentiation of Bergmann glia. Represses neuronal and myogenic differentiation. May play an essential role in postimplantation development, probably in some aspect of cell specification and/or differentiation. May be involved in mesoderm development, somite formation and neurogenesis. May enhance HIF1A function by sequestering HIF1AN away from HIF1A (By similarity). Required for the THBS4 function in regulating protective astrogenesis from the subventricular zone (SVZ) niche after injury. Involved in determination of left/right symmetry by modulating the balance between motile and immotile (sensory) cilia at the left-right organiser (LRO) (By similarity).
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs. Involved in angiogenesis; negatively regulates endothelial cell proliferation and migration and angiogenic sprouting. Involved in the maturation of both CD4(+) and CD8(+) cells in the thymus. Important for follicular differentiation and possibly cell fate selection within the follicle. During cerebellar development, functions as a receptor for neuronal DNER and is involved in the differentiation of Bergmann glia. Represses neuronal and myogenic differentiation. May play an essential role in postimplantation development, probably in some aspect of cell specification and/or differentiation. May be involved in mesoderm development, somite formation and neurogenesis (By similarity). Involved in determination of left/right symmetry by modulating the balance between motile and immotile (sensory) cilia at the left-right organiser (LRO).
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination (By similarity). Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus (By similarity). Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity). Involved in angiogenesis; negatively regulates endothelial cell proliferation and migration and angiogenic sprouting (By similarity). Involved in the maturation of both CD4(+) and CD8(+) cells in the thymus (By similarity). Important for follicular differentiation and possibly cell fate selection within the follicle (By similarity). During cerebellar development, functions as a receptor for neuronal DNER and is involved in the differentiation of Bergmann glia (PubMed:11182080). Represses neuronal and myogenic differentiation (By similarity). May play an essential role in postimplantation development, probably in some aspect of cell specification and/or differentiation (By similarity). May be involved in mesoderm development, somite formation and neurogenesis (By similarity). May enhance HIF1A function by sequestering HIF1AN away from HIF1A (By similarity). Required for the THBS4 function in regulating protective astrogenesis from the subventricular zone (SVZ) niche after injury (By similarity). Involved in determination of left/right symmetry by modulating the balance between motile and immotile (sensory) cilia at the left-right organiser (LRO) (By similarity).
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus (PubMed:21378985, PubMed:21378989). Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity). Involved in bone remodeling and homeostasis. In collaboration with RELA/p65 enhances NFATc1 promoter activity and positively regulates RANKL-induced osteoclast differentiation (PubMed:29149593). Positively regulates self-renewal of liver cancer cells (PubMed:25985737).
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination (PubMed:10393120). Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus (PubMed:10393120, PubMed:18710934). Affects the implementation of differentiation, proliferation and apoptotic programs (PubMed:10393120, PubMed:18710934). May play an essential role in postimplantation development, probably in some aspect of cell specification and/or differentiation (By similarity). In collaboration with RELA/p65 enhances NFATc1 promoter activity and positively regulates RANKL-induced osteoclast differentiation (PubMed:18710934). Positively regulates self-renewal of liver cancer cells (By similarity).
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity). Involved in bone remodeling and homeostasis. In collaboration with RELA/p65 enhances NFATc1 promoter activity and positively regulates RANKL-induced osteoclast differentiation. Positively regulates self-renewal of liver cancer cells (By similarity).
Functions as a receptor for membrane-bound ligands Jagged1, Jagged2 and Delta1 to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity). May play a role during CNS development.
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs. Involved in angiogenesis; negatively regulates endothelial cell proliferation and migration and angiogenic sprouting. Involved in the maturation of both CD4(+) and CD8(+) cells in the thymus. Important for follicular differentiation and possibly cell fate selection within the follicle. During cerebellar development, functions as a receptor for neuronal DNER and is involved in the differentiation of Bergmann glia. Represses neuronal and myogenic differentiation. May play an essential role in postimplantation development, probably in some aspect of cell specification and/or differentiation. May be involved in mesoderm development, somite formation and neurogenesis (By similarity). Involved in determination of left/right symmetry by modulating the balance between motile and immotile (sensory) cilia at the left-right organiser (LRO) (By similarity).
Essential signaling protein which has a major role in many developmental processes (PubMed:3935325). Functions as a receptor for membrane-bound ligands Delta and Serrate to regulate cell-fate determination (PubMed:10935637, PubMed:15620650, PubMed:12909620, PubMed:18243100). Upon ligand activation, and releasing from the cell membrane, the Notch intracellular domain (NICD) forms a transcriptional activator complex with Su(H) (Suppressor of hairless) and activates genes of the E(spl) complex (PubMed:7671825). Regulates oogenesis, the differentiation of the ectoderm and the development of the central and peripheral nervous system, eye, wing disk, muscles and segmental appendages such as antennae and legs, through lateral inhibition or induction (PubMed:11719214, PubMed:12369105, PubMed:3935325). Regulates neuroblast self-renewal, identity and proliferation through the regulation of bHLH-O proteins; in larval brains, involved in the maintenance of type II neuroblast self-renewal and identity by suppressing erm expression together with pnt; might also regulate dpn expression through the activation of the transcriptional regulator Su(H) (PubMed:27151950, PubMed:28899667, PubMed:20152183, PubMed:18342578, PubMed:23056424, PubMed:21262215).
Functions as a receptor for membrane-bound ligands Jagged1, Jagged2 and Delta1 to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity). Acts instructively to control the cell fate determination of CNS multipotent progenitor cells, resulting in astroglial induction and neuron/oligodendrocyte suppression.
Functions as a receptor for membrane-bound ligands Jagged1, Jagged2 and Delta1 to regulate cell-fate determination (PubMed:15350543). Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity).
Required to maintain the integrity of photoreceptor cells (PubMed:27737822, PubMed:28378834, PubMed:30052645). Specifically required for normal morphology of the photoreceptor ciliary pocket, and might thus facilitate protein trafficking between the photoreceptor inner and outer segments via the transition zone (PubMed:27737822).
Functions as a receptor for membrane-bound ligands Jagged1, Jagged2 and Delta1 to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs. May regulate branching morphogenesis in the developing vascular system (By similarity).
Functions as a receptor for membrane-bound ligands Jagged1, Jagged2 and Delta1 to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity). May regulate branching morphogenesis in the developing vascular system.
Cell surface receptor that binds to the chondroitin sulfate moiety of glycosaminoglycan chains and promotes cell attachment. Promotes granulocyte chemotaxis, degranulation and adhesion. In macrophages, promotes the release of inflammatory cytokines, including IL8 and TNF. Signals probably through G-proteins.
Plays a central role in cell polarity establishment (PubMed:2344615, PubMed:12900452, PubMed:10102271, PubMed:11740560). Participates in the assembly, positioning and maintenance of adherens junctions via its interaction with the SAC complex (PubMed:11740560, PubMed:12900452, PubMed:10102271, PubMed:11076972). Controls the coalescence of the spots of zonula adherens (ZA) into a adhesive ring around the cells (PubMed:11740560). It may act as a signal (PubMed:2344615). Involved in morphogenesis of the photoreceptor rhabdomere, for positioning and growth of rhabdomere and AJ during the crucial period of photoreceptor extension along the proximodistal axis of the retina (PubMed:12900452). Component of the crb-galla-Xpd (CGX) complex which is essential for proper mitotic chromosome segregation in early embryos (PubMed:25065591). The CGX complex is also required for cell proliferation in developing wing disks (PubMed:25065591). In the CGX complex, acts with galla-1 or galla-2 to recruit Xpd and thus form the functional complex. Together with apn, plays a key role in trachea development at larval stages (PubMed:30645584).
Ligand for multiple Notch receptors and involved in the mediation of Notch signaling. May be involved in cell-fate decisions during hematopoiesis. Seems to be involved in early and late stages of mammalian cardiovascular development. Inhibits myoblast differentiation (By similarity). May regulate fibroblast growth factor-induced angiogenesis.
Functional ligand of integrin alpha-8/beta-1 in kidney development. Regulates the expression of GDNF with integrin alpha-8/beta-1 which is essential for kidney development. May also play a role in the development and function of various tissues, regulating cell adhesion, spreading and survival through the binding of several integrins (By similarity).
Ligand for multiple Notch receptors and involved in the mediation of Notch signaling (PubMed:18660822, PubMed:20437614). May be involved in cell-fate decisions during hematopoiesis (PubMed:9462510). Seems to be involved in early and late stages of mammalian cardiovascular development. Inhibits myoblast differentiation (By similarity). Enhances fibroblast growth factor-induced angiogenesis (in vitro).
Functional ligand of integrin alpha-8/beta-1 in kidney development. Regulates the expression of GDNF with integrin alpha-8/beta-1 which is essential for kidney development. May also play a role in the development and function of various tissues, regulating cell adhesion, spreading and survival through the binding of several integrins.
Subunit
Interacts with BMP2, BMP4, BMP7, BMP10 and GDF5 (PubMed:18339631). Interacts with MFAP2 and MFAP5 (PubMed:15131124). Interacts with ADAMTSL5 (PubMed:23010571). Interacts with MFAP4 (PubMed:26601954).
Interacts with BMP2, BMP4, BMP7, BMP10 and GDF5. Interacts with MFAP2 and MFAP5. Interacts with ADAMTSL5. Interacts with MFAP4.
Interacts with COL16A1 (PubMed:15165854). Interacts with integrin alpha-V/beta-3 (PubMed:15062093). Interacts with ADAMTS10; this interaction promotes microfibril assembly (PubMed:21402694). Interacts with THSD4; this interaction promotes fibril formation (By similarity). Interacts (via N-terminal domain) with FBLN2, FBLN4 and FBLN5 (PubMed:15790312, PubMed:17255108). Interacts with ELN (PubMed:15790312). Forms a ternary complex with ELN and FBLN2 or FBLN5 and a significant interaction with ELN seen only in the presence of FBLN2 or FBLN5 (PubMed:17255108). Interacts (via N-terminal domain) with LTBP2 (via C-terminal domain) in a Ca(+2)-dependent manner (PubMed:17293099). Interacts (via N-terminal domain) with LTBP1 (via C-terminal domain) (PubMed:17293099). Interacts with integrins ITGA5:ITGB1, ITGAV:ITGB3 and ITGAV:ITGB6 (PubMed:17158881, PubMed:12807887). Interacts (via N-terminal domain) with BMP2, BMP4, BMP7, BMP10 and GDF5 (PubMed:18339631). Interacts (via N-terminal domain) with MFAP2 and MFAP5 (PubMed:15131124). Interacts with ADAMTSL5 (PubMed:23010571). Interacts with MFAP4 (PubMed:26601954). Interacts (via N-terminal domain) with TNFSF11 in a Ca(+2)-dependent manner (PubMed:24039232).
Interacts with COL16A1. Interacts with integrin alpha-V/beta-3. Interacts with ADAMTS10; this interaction promotes microfibril assembly (By similarity). Interacts with THSD4; this interaction promotes fibril formation (PubMed:19940141). Interacts (via N-terminal domain) with FBLN2, FBLN4 and FBLN5. Interacts with ELN. Forms a ternary complex with ELN and FBLN2 or FBLN5 and a significant interaction with ELN seen only in the presence of FBLN2 or FBLN5. Interacts (via N-terminal domain) with LTBP2 (via C-terminal domain) in a Ca(+2)-dependent manner. Interacts (via N-terminal domain) with LTBP1 (via C-terminal domain). Interacts with integrins ITGA5:ITGB1, ITGAV:ITGB3 and ITGAV:ITGB6. Interacts (via N-terminal domain) with BMP2, BMP4, BMP7, BMP10 and GDF5. Interacts (via N-terminal domain) with MFAP2 and MFAP5. Interacts with ADAMTSL5. Interacts with MFAP4. Interacts (via N-terminal domain) with TNFSF11 in a Ca(+2)-dependent manner (By similarity).
Interacts with COL16A1. Interacts with integrin alpha-V/beta-3. Interacts with ADAMTS10; this interaction promotes microfibril assembly. Interacts with THSD4; this interaction promotes fibril formation. Interacts (via N-terminal domain) with FBLN2, FBLN4 and FBLN5. Interacts with ELN. Forms a ternary complex with ELN and FBLN2 or FBLN5 and a significant interaction with ELN seen only in the presence of FBLN2 or FBLN5. Interacts (via N-terminal domain) with LTBP2 (via C-terminal domain) in a Ca(+2)-dependent manner. Interacts (via N-terminal domain) with LTBP1 (via C-terminal domain). Interacts with integrins ITGA5:ITGB1, ITGAV:ITGB3 and ITGAV:ITGB6. Interacts (via N-terminal domain) with BMP2, BMP4, BMP7, BMP10 and GDF5. Interacts (via N-terminal domain) with MFAP2 and MFAP5. Interacts with ADAMTSL5. Interacts with MFAP4. Interacts (via N-terminal domain) with TNFSF11 in a Ca(+2)-dependent manner.
Forms part of the large latent transforming growth factor beta precursor complex; removal is essential for activation of complex (PubMed:7798248). Interacts with SDC4 (PubMed:20382221). Interacts (via C-terminal domain) with FBN1 (via N-terminal domain) in a Ca(+2)-dependent manner (PubMed:17293099).
Forms part of the large latent transforming growth factor beta precursor complex; removal is essential for activation of complex. Interacts with SDC4. Interacts (via C-terminal domain) with FBN1 (via N-terminal domain) in a Ca(+2)-dependent manner.
Interacts with TGFB1; associates via disulfide bonds with the Latency-associated peptide chain (LAP) regulatory chain of TGFB1, leading to regulate activation of TGF-beta-1. LTBP1 does not bind directly to TGF-beta-1, the active chain of TGFB1. Interacts (via C-terminal domain) with FBN1 (via N-terminal domain). Interacts with FBN2. Interacts with ADAMTSL2.
Forms part of the large latent transforming growth factor beta precursor complex; removal is essential for activation of complex (PubMed:9660815). Interacts with LTBP1 and TGFB1 (PubMed:9660815).
Interacts with TGFB1; associates via disulfide bonds with the Latency-associated peptide chain (LAP) regulatory chain of TGFB1, leading to regulate activation of TGF-beta-1 (PubMed:10930463, PubMed:2022183, PubMed:8617200, PubMed:15567420, PubMed:8939931, PubMed:22278742). LTBP1 does not bind directly to TGF-beta-1, the active chain of TGFB1 (PubMed:10930463). Interacts (via C-terminal domain) with FBN1 (via N-terminal domain) (PubMed:12429738, PubMed:17293099). Interacts with FBN2 (PubMed:12429738). Interacts with ADAMTSL2 (PubMed:18677313).
Forms part of the large latent transforming growth factor beta precursor complex; removal is essential for activation of complex.
Forms part of the large latent transforming growth factor beta (TGFB1) precursor complex; removal is essential for activation of complex.
Forms a ternary complex with nrarp and rbpj/suh.
Heterodimer of a C-terminal fragment N(TM) and an N-terminal fragment N(EC) which are probably linked by disulfide bonds. Interacts with DNER, DTX1, DTX2 and RBPJ/RBPSUH. Also interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH1. Notch 1 intracellular domain interacts with SNW1; the interaction involves multimerized NOTCH1 NICD and is implicated in a formation of an intermediate preactivation complex which associates with DNA-bound CBF-1/RBPJ. The activated membrane-bound form interacts with AAK1 which promotes NOTCH1 stabilization. Forms a trimeric complex with FBXW7 and SGK1. Interacts with HIF1AN. HIF1AN negatively regulates the function of notch intracellular domain (NICD), accelerating myogenic differentiation. Interacts (via NICD) with SNAI1 (via zinc fingers); the interaction induces SNAI1 degradation via MDM2-mediated ubiquitination and inhibits SNAI1-induced cell invasion. Interacts (via NICD) with MDM2A. Interacts (via NICD) with BCL6; the interaction decreases MAML1 recruitment by NOTCH1 NICD on target genes DNA and inhibits NOTCH1 transcractivation activity. Interacts with THBS4. Interacts (via the EGF-like repeat region) with CCN3 (via CTCK domain) (PubMed:12050162). Interacts (via EGF-like domains) with DLL4 (via N-terminal DSL and MNNL domains) (By similarity). Interacts with ZMIZ1. Interacts (via NICD domain) with MEGF10 (via the cytoplasmic domain) (PubMed:28498977). Interacts with DLL1 and JAG1 (PubMed:28089369). Interacts (via NICD domain) with PRAG1 (PubMed:25038227). Forms a complex with PRAG1, N1ICD and MAML1, in a MAML1-dependent manner (PubMed:25038227). Interacts (via transmembrane region) with PSEN1; the interaction is direct (By similarity).
Heterodimer of a C-terminal fragment N(TM) and an N-terminal fragment N(EC) which are probably linked by disulfide bonds. Interacts with DNER, DTX1, DTX2 and RBPJ/RBPSUH. Also interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH1. Notch 1 intracellular domain interacts with SNW1; the interaction involves multimerized NOTCH1 NICD and is implicated in a formation of an intermediate preactivation complex which associates with DNA-bound CBF-1/RBPJ. The activated membrane-bound form interacts with AAK1 which promotes NOTCH1 stabilization. Forms a trimeric complex with FBXW7 and SGK1. Interacts with HIF1AN. HIF1AN negatively regulates the function of notch intracellular domain (NICD), accelerating myogenic differentiation. Interacts (via NICD) with SNAI1 (via zinc fingers); the interaction induces SNAI1 degradation via MDM2-mediated ubiquitination and inhibits SNAI1-induced cell invasion. Interacts (via NICD) with MDM2A. Interacts (via NICD) with BCL6; the interaction decreases MAML1 recruitment by NOTCH1 NICD on target genes DNA and inhibits NOTCH1 transcractivation activity (By similarity). Interacts with THBS4 (By similarity). Interacts (via the EGF-like repeat region) with CCN3 (via CTCK domain). Interacts (via EGF-like domains) with DLL4 (via N-terminal DSL and MNNL domains). Interacts with ZMIZ1. Interacts (via NICD domain) with MEGF10 (via the cytoplasmic domain). Interacts with DLL1 and JAG1 (By similarity). Interacts (via NICD domain) with PRAG1 (By similarity). Forms a complex with PRAG1, N1ICD and MAML1, in a MAML1-dependent manner (By similarity). Interacts (via transmembrane region) with PSEN1; the interaction is direct (By similarity).
Heterodimer of a C-terminal fragment N(TM) and an N-terminal fragment N(EC) which are probably linked by disulfide bonds. Interacts with DNER, DTX1, DTX2 and RBPJ/RBPSUH. Also interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH1. Notch 1 intracellular domain interacts with SNW1; the interaction involves multimerized NOTCH1 NICD and is implicated in a formation of an intermediate preactivation complex which associates with DNA-bound CBF-1/RBPJ. The activated membrane-bound form interacts with AAK1 which promotes NOTCH1 stabilization. Forms a trimeric complex with FBXW7 and SGK1. Interacts with HIF1AN. HIF1AN negatively regulates the function of notch intracellular domain (NICD), accelerating myogenic differentiation. Interacts (via NICD) with SNAI1 (via zinc fingers); the interaction induces SNAI1 degradation via MDM2-mediated ubiquitination and inhibits SNAI1-induced cell invasion. Interacts (via NICD) with MDM2A. Interacts (via NICD) with BCL6; the interaction decreases MAML1 recruitment by NOTCH1 NICD on target genes DNA and inhibits NOTCH1 transcractivation activity (By similarity). Interacts with THBS4 (By similarity). Interacts (via the EGF-like repeat region) with CCN3 (via CTCK domain) (By similarity). Interacts (via EGF-like domains) with DLL4 (via N-terminal DSL and MNNL domains) (PubMed:25700513). Interacts with ZMIZ1 (By similarity). Interacts (via NICD domain) with MEGF10 (via the cytoplasmic domain). Interacts with DLL1 and JAG1 (By similarity). Interacts (via NICD domain) with PRAG1 (By similarity). Forms a complex with PRAG1, N1ICD and MAML1, in a MAML1-dependent manner (By similarity). Interacts (via transmembrane region) with PSEN1; the interaction is direct (By similarity).
Heterodimer of a C-terminal fragment N(TM) and an N-terminal fragment N(EC) which are probably linked by disulfide bonds. Interacts with DNER, DTX1, DTX2 and RBPJ/RBPSUH. Also interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH1 (PubMed:11101851, PubMed:12370315). The NOTCH1 intracellular domain interacts with SNW1; the interaction involves multimerized NOTCH1 NICD and is implicated in a formation of an intermediate preactivation complex which associates with DNA-bound CBF-1/RBPJ (PubMed:10713164). The activated membrane-bound form interacts with AAK1 which promotes NOTCH1 stabilization. Forms a trimeric complex with FBXW7 and SGK1. Interacts with HIF1AN. HIF1AN negatively regulates the function of notch intracellular domain (NICD), accelerating myogenic differentiation (PubMed:17573339). Interacts (via NICD) with SNAI1 (via zinc fingers); the interaction induces SNAI1 degradation via MDM2-mediated ubiquitination and inhibits SNAI1-induced cell invasion. Interacts (via NICD) with MDM2A. Interacts (via NICD) with BCL6; the interaction decreases MAML1 recruitment by NOTCH1 NICD on target genes DNA and inhibits NOTCH1 transcractivation activity. Interacts with THBS4 (By similarity). Interacts (via the EGF-like repeat region) with CCN3 (via CTCK domain) (PubMed:12050162). Interacts (via EGF-like domains) with DLL4 (via N-terminal DSL and MNNL domains) (By similarity). Interacts with ZMIZ1. Interacts (via NICD domain) with MEGF10 (via the cytoplasmic domain). Interacts with DLL1 and JAG1 (By similarity). Interacts (via NICD domain) with PRAG1 (By similarity). Forms a complex with PRAG1, N1ICD and MAML1, in a MAML1-dependent manner (By similarity). Interacts (via transmembrane region) with PSEN1; the interaction is direct (PubMed:30598546).
Heterodimer of a C-terminal fragment N(TM) and an N-terminal fragment N(EC) which are probably linked by disulfide bonds (By similarity). Interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH2. Interacts with RELA/p65 (By similarity). Interacts with HIF1AN. Interacts (via ANK repeats) with TCIM, the interaction inhibits the nuclear translocation of NOTCH2 N2ICD (PubMed:25985737). Interacts with CUL1, RBX1, SKP1 and FBXW7 that are SCF(FBXW7) E3 ubiquitin-protein ligase complex components (PubMed:29149593). Interacts with MINAR1; this interaction increases MINAR1 stability and function (PubMed:29329397). Interacts with NOTCH2NL (NOTCH2NLA, NOTCH2NLB and/or NOTCH2NLC); leading to enhance Notch signaling pathway in a non-cell-autonomous manner (PubMed:29856954). Interacts with MDK; this interaction mediates a nuclear accumulation of NOTCH2 and therefore activation of NOTCH2 signaling leading to interaction between HES1 and STAT3 (PubMed:18469519).
Heterodimer of a C-terminal fragment N(TM) and an N-terminal fragment N(EC) which are probably linked by disulfide bonds. Interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH2. Interacts with RELA/p65. Interacts with HIF1AN. Interacts (via ANK repeats) with TCIM, the interaction inhibits the nuclear translocation of NOTCH2 N2ICD (By similarity). Interacts with CUL1, RBX1, SKP1 and FBXW7 that are SCF(FBXW7) E3 ubiquitin-protein ligase complex components. Interacts with MINAR1; this interaction increases MINAR1 stability and function (By similarity). Interacts with MDK; this interaction mediates a nuclear accumulation of NOTCH2 and therefore activation of NOTCH2 signaling leading to interaction between HES1 and STAT3 (By similarity).
Heterodimer of a C-terminal fragment N(TM) and an N-terminal fragment N(EC) which are probably linked by disulfide bonds (By similarity). Interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH2. Interacts with RELA/p65 (By similarity). Interacts with HIF1AN. Interacts (via ANK repeats) with TCIM, the interaction inhibits the nuclear translocation of NOTCH2 N2ICD (By similarity). Interacts with CUL1, RBX1, SKP1 and FBXW7 that are SCF(FBXW7) E3 ubiquitin-protein ligase complex components.Interacts with MINAR1; this interaction increases MINAR1 stability and function (By similarity). Interacts with MDK; this interaction mediates a nuclear accumulation of NOTCH2 and therefore activation of NOTCH2 signaling leading to interaction between HES1 and STAT3 (By similarity).
Interacts with PSMA1 (By similarity). Heterodimer of a C-terminal fragment N(TM) and a N-terminal fragment N(EC) which are probably linked by disulfide bonds. Interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH3. Interacts with HIF1AN.
Homomer. Interacts with Su(H) when activated. Interacts with Dx via its ANK repeats. Interacts with Dl via the EGF repeats and the Dl EGF repeats. Interacts with Nedd4 and Su(dx). Interacts with O-fut1; the interaction glycosylates N and transports N to early endosomes. Interacts with Akap200; the interaction stabilizes N/Notch protein levels by preventing Cbl-mediated ubiquitination and subsequent lysosomal degradation of N/Notch (PubMed:29309414).
Heterodimer of a C-terminal fragment N(TM) and a N-terminal fragment N(EC) which are probably linked by disulfide bonds. Interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH3. Interacts with PSMA1 (By similarity). Interacts with HIF1AN (By similarity).
Heterodimer of a C-terminal fragment N(TM) and a N-terminal fragment N(EC) which are probably linked by disulfide bonds (By similarity). Interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH3. Interacts with PSMA1. Interacts with HIF1AN.
Homotetramer.
Heterodimer of a C-terminal fragment N(TM) and a N-terminal fragment N(EC) which are probably linked by disulfide bonds (By similarity). Interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH4.
Heterodimer of a C-terminal fragment N(TM) and a N-terminal fragment N(EC) which are probably linked by disulfide bonds. Interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH4.
Forms a heterodimer, consisting of a large extracellular region non-covalently linked to a seven-transmembrane moiety. Interacts with chondroitin sulfate; the interaction with chondroitin sulfate is calcium-dependent. Interacts with CD55.
Component of the SAC complex, a complex composed of crb, Patj and sdt (PubMed:11740560, PubMed:10102271, PubMed:11076972). May interact with the par-6 complex, which is composed of par-6, baz and aPKC, via its interaction with Patj (PubMed:12900452, PubMed:10102271, PubMed:11076972). Interacts with other proteins with Patj and sdt via its short cytoplasmic tail (PubMed:11740560). Component of the CGX complex composed of crb, galla (galla-1 or galla-2) and Xpd (PubMed:25065591). Able to interact independently (via intracellular domain) with galla-1, galla-2 and Xpd (PubMed:25065591). Interacts with apn (PubMed:30645584).
Interacts with NOTCH1, NOTCH2 and NOTCH3.
Homodimer and homotrimer.
Interacts with NOTCH1 (PubMed:28089369). Interacts with NOTCH2 and NOTCH3 (By similarity).
Interacts with NOTCH2 and NOTCH3 (By similarity). Interacts with NOTCH1 (in the presence of calcium ions) (PubMed:18660822).
Interacts with BMP2, BMP4, BMP7, BMP10 and GDF5. Interacts with MFAP2 and MFAP5. Interacts with ADAMTSL5. Interacts with MFAP4.
Interacts with COL16A1 (PubMed:15165854). Interacts with integrin alpha-V/beta-3 (PubMed:15062093). Interacts with ADAMTS10; this interaction promotes microfibril assembly (PubMed:21402694). Interacts with THSD4; this interaction promotes fibril formation (By similarity). Interacts (via N-terminal domain) with FBLN2, FBLN4 and FBLN5 (PubMed:15790312, PubMed:17255108). Interacts with ELN (PubMed:15790312). Forms a ternary complex with ELN and FBLN2 or FBLN5 and a significant interaction with ELN seen only in the presence of FBLN2 or FBLN5 (PubMed:17255108). Interacts (via N-terminal domain) with LTBP2 (via C-terminal domain) in a Ca(+2)-dependent manner (PubMed:17293099). Interacts (via N-terminal domain) with LTBP1 (via C-terminal domain) (PubMed:17293099). Interacts with integrins ITGA5:ITGB1, ITGAV:ITGB3 and ITGAV:ITGB6 (PubMed:17158881, PubMed:12807887). Interacts (via N-terminal domain) with BMP2, BMP4, BMP7, BMP10 and GDF5 (PubMed:18339631). Interacts (via N-terminal domain) with MFAP2 and MFAP5 (PubMed:15131124). Interacts with ADAMTSL5 (PubMed:23010571). Interacts with MFAP4 (PubMed:26601954). Interacts (via N-terminal domain) with TNFSF11 in a Ca(+2)-dependent manner (PubMed:24039232).
Interacts with COL16A1. Interacts with integrin alpha-V/beta-3. Interacts with ADAMTS10; this interaction promotes microfibril assembly (By similarity). Interacts with THSD4; this interaction promotes fibril formation (PubMed:19940141). Interacts (via N-terminal domain) with FBLN2, FBLN4 and FBLN5. Interacts with ELN. Forms a ternary complex with ELN and FBLN2 or FBLN5 and a significant interaction with ELN seen only in the presence of FBLN2 or FBLN5. Interacts (via N-terminal domain) with LTBP2 (via C-terminal domain) in a Ca(+2)-dependent manner. Interacts (via N-terminal domain) with LTBP1 (via C-terminal domain). Interacts with integrins ITGA5:ITGB1, ITGAV:ITGB3 and ITGAV:ITGB6. Interacts (via N-terminal domain) with BMP2, BMP4, BMP7, BMP10 and GDF5. Interacts (via N-terminal domain) with MFAP2 and MFAP5. Interacts with ADAMTSL5. Interacts with MFAP4. Interacts (via N-terminal domain) with TNFSF11 in a Ca(+2)-dependent manner (By similarity).
Interacts with COL16A1. Interacts with integrin alpha-V/beta-3. Interacts with ADAMTS10; this interaction promotes microfibril assembly. Interacts with THSD4; this interaction promotes fibril formation. Interacts (via N-terminal domain) with FBLN2, FBLN4 and FBLN5. Interacts with ELN. Forms a ternary complex with ELN and FBLN2 or FBLN5 and a significant interaction with ELN seen only in the presence of FBLN2 or FBLN5. Interacts (via N-terminal domain) with LTBP2 (via C-terminal domain) in a Ca(+2)-dependent manner. Interacts (via N-terminal domain) with LTBP1 (via C-terminal domain). Interacts with integrins ITGA5:ITGB1, ITGAV:ITGB3 and ITGAV:ITGB6. Interacts (via N-terminal domain) with BMP2, BMP4, BMP7, BMP10 and GDF5. Interacts (via N-terminal domain) with MFAP2 and MFAP5. Interacts with ADAMTSL5. Interacts with MFAP4. Interacts (via N-terminal domain) with TNFSF11 in a Ca(+2)-dependent manner.
Forms part of the large latent transforming growth factor beta precursor complex; removal is essential for activation of complex (PubMed:7798248). Interacts with SDC4 (PubMed:20382221). Interacts (via C-terminal domain) with FBN1 (via N-terminal domain) in a Ca(+2)-dependent manner (PubMed:17293099).
Forms part of the large latent transforming growth factor beta precursor complex; removal is essential for activation of complex. Interacts with SDC4. Interacts (via C-terminal domain) with FBN1 (via N-terminal domain) in a Ca(+2)-dependent manner.
Interacts with TGFB1; associates via disulfide bonds with the Latency-associated peptide chain (LAP) regulatory chain of TGFB1, leading to regulate activation of TGF-beta-1. LTBP1 does not bind directly to TGF-beta-1, the active chain of TGFB1. Interacts (via C-terminal domain) with FBN1 (via N-terminal domain). Interacts with FBN2. Interacts with ADAMTSL2.
Forms part of the large latent transforming growth factor beta precursor complex; removal is essential for activation of complex (PubMed:9660815). Interacts with LTBP1 and TGFB1 (PubMed:9660815).
Interacts with TGFB1; associates via disulfide bonds with the Latency-associated peptide chain (LAP) regulatory chain of TGFB1, leading to regulate activation of TGF-beta-1 (PubMed:10930463, PubMed:2022183, PubMed:8617200, PubMed:15567420, PubMed:8939931, PubMed:22278742). LTBP1 does not bind directly to TGF-beta-1, the active chain of TGFB1 (PubMed:10930463). Interacts (via C-terminal domain) with FBN1 (via N-terminal domain) (PubMed:12429738, PubMed:17293099). Interacts with FBN2 (PubMed:12429738). Interacts with ADAMTSL2 (PubMed:18677313).
Forms part of the large latent transforming growth factor beta precursor complex; removal is essential for activation of complex.
Forms part of the large latent transforming growth factor beta (TGFB1) precursor complex; removal is essential for activation of complex.
Forms a ternary complex with nrarp and rbpj/suh.
Heterodimer of a C-terminal fragment N(TM) and an N-terminal fragment N(EC) which are probably linked by disulfide bonds. Interacts with DNER, DTX1, DTX2 and RBPJ/RBPSUH. Also interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH1. Notch 1 intracellular domain interacts with SNW1; the interaction involves multimerized NOTCH1 NICD and is implicated in a formation of an intermediate preactivation complex which associates with DNA-bound CBF-1/RBPJ. The activated membrane-bound form interacts with AAK1 which promotes NOTCH1 stabilization. Forms a trimeric complex with FBXW7 and SGK1. Interacts with HIF1AN. HIF1AN negatively regulates the function of notch intracellular domain (NICD), accelerating myogenic differentiation. Interacts (via NICD) with SNAI1 (via zinc fingers); the interaction induces SNAI1 degradation via MDM2-mediated ubiquitination and inhibits SNAI1-induced cell invasion. Interacts (via NICD) with MDM2A. Interacts (via NICD) with BCL6; the interaction decreases MAML1 recruitment by NOTCH1 NICD on target genes DNA and inhibits NOTCH1 transcractivation activity. Interacts with THBS4. Interacts (via the EGF-like repeat region) with CCN3 (via CTCK domain) (PubMed:12050162). Interacts (via EGF-like domains) with DLL4 (via N-terminal DSL and MNNL domains) (By similarity). Interacts with ZMIZ1. Interacts (via NICD domain) with MEGF10 (via the cytoplasmic domain) (PubMed:28498977). Interacts with DLL1 and JAG1 (PubMed:28089369). Interacts (via NICD domain) with PRAG1 (PubMed:25038227). Forms a complex with PRAG1, N1ICD and MAML1, in a MAML1-dependent manner (PubMed:25038227). Interacts (via transmembrane region) with PSEN1; the interaction is direct (By similarity).
Heterodimer of a C-terminal fragment N(TM) and an N-terminal fragment N(EC) which are probably linked by disulfide bonds. Interacts with DNER, DTX1, DTX2 and RBPJ/RBPSUH. Also interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH1. Notch 1 intracellular domain interacts with SNW1; the interaction involves multimerized NOTCH1 NICD and is implicated in a formation of an intermediate preactivation complex which associates with DNA-bound CBF-1/RBPJ. The activated membrane-bound form interacts with AAK1 which promotes NOTCH1 stabilization. Forms a trimeric complex with FBXW7 and SGK1. Interacts with HIF1AN. HIF1AN negatively regulates the function of notch intracellular domain (NICD), accelerating myogenic differentiation. Interacts (via NICD) with SNAI1 (via zinc fingers); the interaction induces SNAI1 degradation via MDM2-mediated ubiquitination and inhibits SNAI1-induced cell invasion. Interacts (via NICD) with MDM2A. Interacts (via NICD) with BCL6; the interaction decreases MAML1 recruitment by NOTCH1 NICD on target genes DNA and inhibits NOTCH1 transcractivation activity (By similarity). Interacts with THBS4 (By similarity). Interacts (via the EGF-like repeat region) with CCN3 (via CTCK domain). Interacts (via EGF-like domains) with DLL4 (via N-terminal DSL and MNNL domains). Interacts with ZMIZ1. Interacts (via NICD domain) with MEGF10 (via the cytoplasmic domain). Interacts with DLL1 and JAG1 (By similarity). Interacts (via NICD domain) with PRAG1 (By similarity). Forms a complex with PRAG1, N1ICD and MAML1, in a MAML1-dependent manner (By similarity). Interacts (via transmembrane region) with PSEN1; the interaction is direct (By similarity).
Heterodimer of a C-terminal fragment N(TM) and an N-terminal fragment N(EC) which are probably linked by disulfide bonds. Interacts with DNER, DTX1, DTX2 and RBPJ/RBPSUH. Also interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH1. Notch 1 intracellular domain interacts with SNW1; the interaction involves multimerized NOTCH1 NICD and is implicated in a formation of an intermediate preactivation complex which associates with DNA-bound CBF-1/RBPJ. The activated membrane-bound form interacts with AAK1 which promotes NOTCH1 stabilization. Forms a trimeric complex with FBXW7 and SGK1. Interacts with HIF1AN. HIF1AN negatively regulates the function of notch intracellular domain (NICD), accelerating myogenic differentiation. Interacts (via NICD) with SNAI1 (via zinc fingers); the interaction induces SNAI1 degradation via MDM2-mediated ubiquitination and inhibits SNAI1-induced cell invasion. Interacts (via NICD) with MDM2A. Interacts (via NICD) with BCL6; the interaction decreases MAML1 recruitment by NOTCH1 NICD on target genes DNA and inhibits NOTCH1 transcractivation activity (By similarity). Interacts with THBS4 (By similarity). Interacts (via the EGF-like repeat region) with CCN3 (via CTCK domain) (By similarity). Interacts (via EGF-like domains) with DLL4 (via N-terminal DSL and MNNL domains) (PubMed:25700513). Interacts with ZMIZ1 (By similarity). Interacts (via NICD domain) with MEGF10 (via the cytoplasmic domain). Interacts with DLL1 and JAG1 (By similarity). Interacts (via NICD domain) with PRAG1 (By similarity). Forms a complex with PRAG1, N1ICD and MAML1, in a MAML1-dependent manner (By similarity). Interacts (via transmembrane region) with PSEN1; the interaction is direct (By similarity).
Heterodimer of a C-terminal fragment N(TM) and an N-terminal fragment N(EC) which are probably linked by disulfide bonds. Interacts with DNER, DTX1, DTX2 and RBPJ/RBPSUH. Also interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH1 (PubMed:11101851, PubMed:12370315). The NOTCH1 intracellular domain interacts with SNW1; the interaction involves multimerized NOTCH1 NICD and is implicated in a formation of an intermediate preactivation complex which associates with DNA-bound CBF-1/RBPJ (PubMed:10713164). The activated membrane-bound form interacts with AAK1 which promotes NOTCH1 stabilization. Forms a trimeric complex with FBXW7 and SGK1. Interacts with HIF1AN. HIF1AN negatively regulates the function of notch intracellular domain (NICD), accelerating myogenic differentiation (PubMed:17573339). Interacts (via NICD) with SNAI1 (via zinc fingers); the interaction induces SNAI1 degradation via MDM2-mediated ubiquitination and inhibits SNAI1-induced cell invasion. Interacts (via NICD) with MDM2A. Interacts (via NICD) with BCL6; the interaction decreases MAML1 recruitment by NOTCH1 NICD on target genes DNA and inhibits NOTCH1 transcractivation activity. Interacts with THBS4 (By similarity). Interacts (via the EGF-like repeat region) with CCN3 (via CTCK domain) (PubMed:12050162). Interacts (via EGF-like domains) with DLL4 (via N-terminal DSL and MNNL domains) (By similarity). Interacts with ZMIZ1. Interacts (via NICD domain) with MEGF10 (via the cytoplasmic domain). Interacts with DLL1 and JAG1 (By similarity). Interacts (via NICD domain) with PRAG1 (By similarity). Forms a complex with PRAG1, N1ICD and MAML1, in a MAML1-dependent manner (By similarity). Interacts (via transmembrane region) with PSEN1; the interaction is direct (PubMed:30598546).
Heterodimer of a C-terminal fragment N(TM) and an N-terminal fragment N(EC) which are probably linked by disulfide bonds (By similarity). Interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH2. Interacts with RELA/p65 (By similarity). Interacts with HIF1AN. Interacts (via ANK repeats) with TCIM, the interaction inhibits the nuclear translocation of NOTCH2 N2ICD (PubMed:25985737). Interacts with CUL1, RBX1, SKP1 and FBXW7 that are SCF(FBXW7) E3 ubiquitin-protein ligase complex components (PubMed:29149593). Interacts with MINAR1; this interaction increases MINAR1 stability and function (PubMed:29329397). Interacts with NOTCH2NL (NOTCH2NLA, NOTCH2NLB and/or NOTCH2NLC); leading to enhance Notch signaling pathway in a non-cell-autonomous manner (PubMed:29856954). Interacts with MDK; this interaction mediates a nuclear accumulation of NOTCH2 and therefore activation of NOTCH2 signaling leading to interaction between HES1 and STAT3 (PubMed:18469519).
Heterodimer of a C-terminal fragment N(TM) and an N-terminal fragment N(EC) which are probably linked by disulfide bonds. Interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH2. Interacts with RELA/p65. Interacts with HIF1AN. Interacts (via ANK repeats) with TCIM, the interaction inhibits the nuclear translocation of NOTCH2 N2ICD (By similarity). Interacts with CUL1, RBX1, SKP1 and FBXW7 that are SCF(FBXW7) E3 ubiquitin-protein ligase complex components. Interacts with MINAR1; this interaction increases MINAR1 stability and function (By similarity). Interacts with MDK; this interaction mediates a nuclear accumulation of NOTCH2 and therefore activation of NOTCH2 signaling leading to interaction between HES1 and STAT3 (By similarity).
Heterodimer of a C-terminal fragment N(TM) and an N-terminal fragment N(EC) which are probably linked by disulfide bonds (By similarity). Interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH2. Interacts with RELA/p65 (By similarity). Interacts with HIF1AN. Interacts (via ANK repeats) with TCIM, the interaction inhibits the nuclear translocation of NOTCH2 N2ICD (By similarity). Interacts with CUL1, RBX1, SKP1 and FBXW7 that are SCF(FBXW7) E3 ubiquitin-protein ligase complex components.Interacts with MINAR1; this interaction increases MINAR1 stability and function (By similarity). Interacts with MDK; this interaction mediates a nuclear accumulation of NOTCH2 and therefore activation of NOTCH2 signaling leading to interaction between HES1 and STAT3 (By similarity).
Interacts with PSMA1 (By similarity). Heterodimer of a C-terminal fragment N(TM) and a N-terminal fragment N(EC) which are probably linked by disulfide bonds. Interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH3. Interacts with HIF1AN.
Homomer. Interacts with Su(H) when activated. Interacts with Dx via its ANK repeats. Interacts with Dl via the EGF repeats and the Dl EGF repeats. Interacts with Nedd4 and Su(dx). Interacts with O-fut1; the interaction glycosylates N and transports N to early endosomes. Interacts with Akap200; the interaction stabilizes N/Notch protein levels by preventing Cbl-mediated ubiquitination and subsequent lysosomal degradation of N/Notch (PubMed:29309414).
Heterodimer of a C-terminal fragment N(TM) and a N-terminal fragment N(EC) which are probably linked by disulfide bonds. Interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH3. Interacts with PSMA1 (By similarity). Interacts with HIF1AN (By similarity).
Heterodimer of a C-terminal fragment N(TM) and a N-terminal fragment N(EC) which are probably linked by disulfide bonds (By similarity). Interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH3. Interacts with PSMA1. Interacts with HIF1AN.
Homotetramer.
Heterodimer of a C-terminal fragment N(TM) and a N-terminal fragment N(EC) which are probably linked by disulfide bonds (By similarity). Interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH4.
Heterodimer of a C-terminal fragment N(TM) and a N-terminal fragment N(EC) which are probably linked by disulfide bonds. Interacts with MAML1, MAML2 and MAML3 which act as transcriptional coactivators for NOTCH4.
Forms a heterodimer, consisting of a large extracellular region non-covalently linked to a seven-transmembrane moiety. Interacts with chondroitin sulfate; the interaction with chondroitin sulfate is calcium-dependent. Interacts with CD55.
Component of the SAC complex, a complex composed of crb, Patj and sdt (PubMed:11740560, PubMed:10102271, PubMed:11076972). May interact with the par-6 complex, which is composed of par-6, baz and aPKC, via its interaction with Patj (PubMed:12900452, PubMed:10102271, PubMed:11076972). Interacts with other proteins with Patj and sdt via its short cytoplasmic tail (PubMed:11740560). Component of the CGX complex composed of crb, galla (galla-1 or galla-2) and Xpd (PubMed:25065591). Able to interact independently (via intracellular domain) with galla-1, galla-2 and Xpd (PubMed:25065591). Interacts with apn (PubMed:30645584).
Interacts with NOTCH1, NOTCH2 and NOTCH3.
Homodimer and homotrimer.
Interacts with NOTCH1 (PubMed:28089369). Interacts with NOTCH2 and NOTCH3 (By similarity).
Interacts with NOTCH2 and NOTCH3 (By similarity). Interacts with NOTCH1 (in the presence of calcium ions) (PubMed:18660822).
Miscellaneous
Asprosin: Was named after the Greek word for white, because of the reduction in subcutaneous white adipose tissue that is displayed by asprosin-deficient patients.
Was named nephronectin based on its potential role in kidney development.
Was named nephronectin based on its potential role in kidney development.
Similarity
Belongs to the fibrillin family.
Belongs to the LTBP family.
Belongs to the NOTCH family.
Belongs to the EYS family.
Belongs to the G-protein coupled receptor 2 family. Adhesion G-protein coupled receptor (ADGR) subfamily.
Belongs to the Crumbs protein family.
Belongs to the nephronectin family.
Belongs to the LTBP family.
Belongs to the NOTCH family.
Belongs to the EYS family.
Belongs to the G-protein coupled receptor 2 family. Adhesion G-protein coupled receptor (ADGR) subfamily.
Belongs to the Crumbs protein family.
Belongs to the nephronectin family.
Keywords
Alternative splicing
Calcium
Complete proteome
Disease mutation
Disulfide bond
EGF-like domain
Extracellular matrix
Glycoprotein
Polymorphism
Reference proteome
Repeat
Secreted
Signal
Direct protein sequencing
3D-structure
Aortic aneurysm
Dwarfism
Heparin-binding
Hormone
Phosphoprotein
Glaucoma
Growth factor binding
Hydroxylation
Cleavage on pair of basic residues
Amelogenesis imperfecta
Activator
Angiogenesis
ANK repeat
Cell membrane
Developmental protein
Differentiation
Membrane
Metal-binding
Notch signaling pathway
Nucleus
Receptor
Transcription
Transcription regulation
Transmembrane
Transmembrane helix
Isopeptide bond
Ubl conjugation
Cytoplasm
Methylation
Endosome
Neurogenesis
Oogenesis
Cytoplasmic vesicle
Cell projection
Cytoskeleton
Sensory transduction
Vision
Triplet repeat expansion
Proto-oncogene
Autocatalytic cleavage
Cell adhesion
Inflammatory response
Cell cycle
Cell division
Mitosis
Deafness
Feature
chain Fibrillin-2
splice variant In isoform 2.
sequence variant In DA9; dbSNP:rs137852826.
glycosylation site N-linked (GlcNAc...) asparagine
splice variant In isoform 2.
sequence variant In DA9; dbSNP:rs137852826.
glycosylation site N-linked (GlcNAc...) asparagine
Uniprot
P35556.3
Q61555.2
P35555.4
Q61554.2
Q9TV36.1
Q75N90.3
+ More
P98133.2 Q14767.3 O35806.1 O08999.2 Q8CG19.2 Q28019.2 Q8N2S1.2 Q00918.1 Q14766.4 Q9NS15.4 Q61810.4 P21783.3 Q01705.3 G3I6Z6.2 A2RUV0.2 Q07008.3 P46531.4 Q04721.3 O35516.2 Q9QW30.1 Q61982.1 P46530.1 P07207.3 Q9R172.2 Q9UM47.2 P10079.2 B8JI71.2 O88281.1 Q80V70.3 Q99466.2 P31695.3 O75095.4 Q2Q421.2 P10040.3 Q9QXX0.1 Q5RBP1.1 Q6UXI9.3 Q63722.2 P78504.3 Q91V88.1
P98133.2 Q14767.3 O35806.1 O08999.2 Q8CG19.2 Q28019.2 Q8N2S1.2 Q00918.1 Q14766.4 Q9NS15.4 Q61810.4 P21783.3 Q01705.3 G3I6Z6.2 A2RUV0.2 Q07008.3 P46531.4 Q04721.3 O35516.2 Q9QW30.1 Q61982.1 P46530.1 P07207.3 Q9R172.2 Q9UM47.2 P10079.2 B8JI71.2 O88281.1 Q80V70.3 Q99466.2 P31695.3 O75095.4 Q2Q421.2 P10040.3 Q9QXX0.1 Q5RBP1.1 Q6UXI9.3 Q63722.2 P78504.3 Q91V88.1
Pubmed
8120105
14702039
15372022
1852206
15131124
18339631
+ More
19159218 23010571 24899048 26601954 7493032 9714438 9737771 10797416 11754102 19006240 20799338 21248752 25834781 27196565 7744963 19468303 8307578 11470817 16407178 20855508 24039232 8364578 15221638 16572171 15489334 7691719 1852207 8430317 10636927 9817919 2739055 1860873 11461921 12807887 15165854 15790312 17158881 17255108 17293099 20979188 21594992 21594993 21402694 24665001 24039054 24613577 24275569 24982166 26091039 27026396 27087445 26860060 9362480 8568869 8653794 12511552 15062093 19446531 24035709 8594563 10633129 12203987 1852208 1301946 1569206 8406497 8504310 8281141 7977366 8188302 8004112 7951214 7911051 8040326 8071963 7870075 8136837 7762551 7611299 7738200 8882780 8863159 9254848 9150726 9338581 9338588 9401003 8988160 9016526 9837823 9452085 10694921 10441597 10425041 11700157 12203992 12402346 11826022 12161601 12525539 14695540 15161917 16222657 16220557 17657824 18435798 19533785 19293843 20803651 21034599 19941982 20547088 20375004 21683322 21542060 22772377 7829516 21183079 19940141 10036187 14962672 11347906 15057824 7835900 19393038 8557636 21989719 7798248 8697098 12508121 10930463 10743502 11104663 19361779 20617341 20382221 22539340 10028916 22673903 26479776 9602168 16141072 12711388 12711389 8524260 9271198 9660815 16157329 19836010 23440719 25641372 2247454 7593177 2350783 15815621 8537398 2022183 8617200 8939931 10677208 12429738 15184403 15567420 18088087 18677313 22278742 14607119 12154076 9620332 17974005 19344874 27068007 7730318 12062452 2402639 11485984 8449489 7956822 12123574 12807718 1426644 8440332 1425352 9653148 10437788 8486742 18299578 9384671 10882062 10882063 11518718 11459941 11226752 12050162 15019995 15240571 15965470 18628966 21464124 21757702 23160044 23886940 23615612 25038227 28089369 28498977 30127001 15802643 17573339 21804562 10734111 20431018 24226769 1764995 15057822 11182080 1295745 11438922 25700513 15164053 1831692 10079256 9590294 10713164 11101851 12370315 16025100 18669648 20616313 22128911 21147854 21245387 25547411 26522984 12795601 15576031 16011479 16530044 28439555 30598546 25132448 16710414 1303260 9244302 17081983 18469519 19690332 20068231 21681853 21378989 21378985 23186163 25985737 29149593 29856954 29329397 17401372 16773578 21793104 21712856 23389697 8917536 10393120 7609614 18710934 19144319 21949356 11577080 7918097 8297791 3935325 3097517 10731132 12537572 10731137 3037327 2981631 2780284 8162848 7671825 10206646 10206647 10935637 11719214 11799064 12369105 12909620 15620649 15620650 17329366 18243100 18342578 18327897 20152183 21262215 23056424 27268051 27151950 28899667 29309414 14573873 8878478 17292860 24129315 25394726 9388399 10227618 10371548 10854111 11058919 11102981 10802807 11559313 11755616 11810186 12146805 12136071 12589106 12810003 15229130 15364702 15378071 15350543 15300988 16009764 15818833 18273901 23731542 24000151 2514273 3498216 2784773 2060714 15659374 23594743 27737822 28378834 30052645 9693030 9168133 9693032 14574404 1312643 9150355 8681805 14656967 10233982 11344305 12168954 2344615 12537569 3107986 10102271 11076972 11740560 12900452 17893096 25065591 30645584 10551863 10556292 12975309 15754038 7697721 9268641 9207788 9462510 10329626 11780052 8955070 9207787 10978356 21269460 18660822 19586525 9585603 10220506 10533065 11058898 11152664 11157803 11139247 11180599 12022040 12297837 12442286 12649809 12497640 15712272 16575836 23801938 20437614 11546798 11470831 17537792
19159218 23010571 24899048 26601954 7493032 9714438 9737771 10797416 11754102 19006240 20799338 21248752 25834781 27196565 7744963 19468303 8307578 11470817 16407178 20855508 24039232 8364578 15221638 16572171 15489334 7691719 1852207 8430317 10636927 9817919 2739055 1860873 11461921 12807887 15165854 15790312 17158881 17255108 17293099 20979188 21594992 21594993 21402694 24665001 24039054 24613577 24275569 24982166 26091039 27026396 27087445 26860060 9362480 8568869 8653794 12511552 15062093 19446531 24035709 8594563 10633129 12203987 1852208 1301946 1569206 8406497 8504310 8281141 7977366 8188302 8004112 7951214 7911051 8040326 8071963 7870075 8136837 7762551 7611299 7738200 8882780 8863159 9254848 9150726 9338581 9338588 9401003 8988160 9016526 9837823 9452085 10694921 10441597 10425041 11700157 12203992 12402346 11826022 12161601 12525539 14695540 15161917 16222657 16220557 17657824 18435798 19533785 19293843 20803651 21034599 19941982 20547088 20375004 21683322 21542060 22772377 7829516 21183079 19940141 10036187 14962672 11347906 15057824 7835900 19393038 8557636 21989719 7798248 8697098 12508121 10930463 10743502 11104663 19361779 20617341 20382221 22539340 10028916 22673903 26479776 9602168 16141072 12711388 12711389 8524260 9271198 9660815 16157329 19836010 23440719 25641372 2247454 7593177 2350783 15815621 8537398 2022183 8617200 8939931 10677208 12429738 15184403 15567420 18088087 18677313 22278742 14607119 12154076 9620332 17974005 19344874 27068007 7730318 12062452 2402639 11485984 8449489 7956822 12123574 12807718 1426644 8440332 1425352 9653148 10437788 8486742 18299578 9384671 10882062 10882063 11518718 11459941 11226752 12050162 15019995 15240571 15965470 18628966 21464124 21757702 23160044 23886940 23615612 25038227 28089369 28498977 30127001 15802643 17573339 21804562 10734111 20431018 24226769 1764995 15057822 11182080 1295745 11438922 25700513 15164053 1831692 10079256 9590294 10713164 11101851 12370315 16025100 18669648 20616313 22128911 21147854 21245387 25547411 26522984 12795601 15576031 16011479 16530044 28439555 30598546 25132448 16710414 1303260 9244302 17081983 18469519 19690332 20068231 21681853 21378989 21378985 23186163 25985737 29149593 29856954 29329397 17401372 16773578 21793104 21712856 23389697 8917536 10393120 7609614 18710934 19144319 21949356 11577080 7918097 8297791 3935325 3097517 10731132 12537572 10731137 3037327 2981631 2780284 8162848 7671825 10206646 10206647 10935637 11719214 11799064 12369105 12909620 15620649 15620650 17329366 18243100 18342578 18327897 20152183 21262215 23056424 27268051 27151950 28899667 29309414 14573873 8878478 17292860 24129315 25394726 9388399 10227618 10371548 10854111 11058919 11102981 10802807 11559313 11755616 11810186 12146805 12136071 12589106 12810003 15229130 15364702 15378071 15350543 15300988 16009764 15818833 18273901 23731542 24000151 2514273 3498216 2784773 2060714 15659374 23594743 27737822 28378834 30052645 9693030 9168133 9693032 14574404 1312643 9150355 8681805 14656967 10233982 11344305 12168954 2344615 12537569 3107986 10102271 11076972 11740560 12900452 17893096 25065591 30645584 10551863 10556292 12975309 15754038 7697721 9268641 9207788 9462510 10329626 11780052 8955070 9207787 10978356 21269460 18660822 19586525 9585603 10220506 10533065 11058898 11152664 11157803 11139247 11180599 12022040 12297837 12442286 12649809 12497640 15712272 16575836 23801938 20437614 11546798 11470831 17537792
EMBL
U03272
AK300440
AC025169
AC034235
AC113387
X62009
+ More
AB209735 L39790 AC102317 AC127358 S69359 L13923 AB177803 GU143398 AC022467 AC084757 AC084758 CH471082 BC146854 L19896 X63556 X62008 S54426 S54425 L29454 U22493 AL844547 AL928930 AF073800 AY165863 AY165864 AY165865 AB177797 AB177798 AB177799 AB177800 AB053450 AC008946 AC022146 L28748 DAAA02029120 DAAA02029121 DAAA02029122 Z37976 S82451 AC013451 Y12760 AF004874 AK052980 AF346465 AF346434 AF346435 AF346436 AF346437 AF346438 AF346439 AF346440 AF346441 AF346442 AF346443 AF346444 AF346445 AF346446 AF346447 AF346448 AF346449 AF346450 AF346451 AF346452 AF346453 AF346454 AF346455 AF346456 AF346457 AF346458 AF346459 AF346460 AF346461 AF346462 AF346463 AF346464 AY143161 AF022889 AC118018 AC126550 AC154178 AC154491 CT033758 BC094612 AK050380 AK080024 AAN38831.1 U35363 Y13622 AF051344 AF051345 AK074499 AC010412 M55431 M34057 AF489528 AB208801 AC019195 AC019127 AC020594 CH471053 BC130289 L48925 BP291349 AF135960 AK024477 AF011407 BC008761 AL117551 L40459 AK080869 AC134563 M33874 Z11886 AF508809 AB100603 AL732541 CH466542 BC138441 BC138442 L02613 X68278 AK154528 AK157475 AJ238029 X82562 JH001398 AAMC01043423 AAMC01043424 AAMC01043425 AAMC01043426 BC133053 X57405 AABR06021907 AABR06021908 CH474001 AF308602 AL592301 AL354671 M73980 AB209873 AF308601 AF315356 AL359752 AL512503 AL596222 U77493 D32210 AC154173 AC164091 CH466620 X68279 U31881 M93661 X74760 X69088 M16152 M16153 M16149 M16150 M16151 K03508 M13689 K03507 AE014298 AL035436 AL035395 M16025 M12175 AF164486 U97669 AF058900 AF058881 AF058882 AF058883 AF058884 AF058885 AF058886 AF058887 AF058888 AF058889 AF058890 AF058891 AF058892 AF058893 AF058894 AF058895 AF058896 AF058897 AF058898 AF058899 AC004257 AC004663 L08692 X17530 M17421 X17533 BX005106 BX323836 CR456624 CU570696 AB011532 AL627123 BC031402 BC039980 BC058571 D63395 D86566 U95299 U89335 AL662884 BX284686 BX284927 CR812478 CR933878 AB023961 AB024520 AB024578 M80456 U43691 AF030001 CT009767 AB016771 AB016772 AB016773 AB016774 AB011539 AB231860 AL512413 AL513320 DQ227276 M33753 AE014297 AY118509 X05144 AF171092 BC058675 CR858597 AY358336 AK290029 AK295772 AK302477 AK304279 AK304721 AC093782 AC109361 CH471057 L38483 AF003837 U73936 AF028593 U61276 AK302554 AL035456 BC126205 BC126207 U77720 AB059656 AF397007 AF397008 AY035898 AY035899 AK050484 BC046642 BC068308
AB209735 L39790 AC102317 AC127358 S69359 L13923 AB177803 GU143398 AC022467 AC084757 AC084758 CH471082 BC146854 L19896 X63556 X62008 S54426 S54425 L29454 U22493 AL844547 AL928930 AF073800 AY165863 AY165864 AY165865 AB177797 AB177798 AB177799 AB177800 AB053450 AC008946 AC022146 L28748 DAAA02029120 DAAA02029121 DAAA02029122 Z37976 S82451 AC013451 Y12760 AF004874 AK052980 AF346465 AF346434 AF346435 AF346436 AF346437 AF346438 AF346439 AF346440 AF346441 AF346442 AF346443 AF346444 AF346445 AF346446 AF346447 AF346448 AF346449 AF346450 AF346451 AF346452 AF346453 AF346454 AF346455 AF346456 AF346457 AF346458 AF346459 AF346460 AF346461 AF346462 AF346463 AF346464 AY143161 AF022889 AC118018 AC126550 AC154178 AC154491 CT033758 BC094612 AK050380 AK080024 AAN38831.1 U35363 Y13622 AF051344 AF051345 AK074499 AC010412 M55431 M34057 AF489528 AB208801 AC019195 AC019127 AC020594 CH471053 BC130289 L48925 BP291349 AF135960 AK024477 AF011407 BC008761 AL117551 L40459 AK080869 AC134563 M33874 Z11886 AF508809 AB100603 AL732541 CH466542 BC138441 BC138442 L02613 X68278 AK154528 AK157475 AJ238029 X82562 JH001398 AAMC01043423 AAMC01043424 AAMC01043425 AAMC01043426 BC133053 X57405 AABR06021907 AABR06021908 CH474001 AF308602 AL592301 AL354671 M73980 AB209873 AF308601 AF315356 AL359752 AL512503 AL596222 U77493 D32210 AC154173 AC164091 CH466620 X68279 U31881 M93661 X74760 X69088 M16152 M16153 M16149 M16150 M16151 K03508 M13689 K03507 AE014298 AL035436 AL035395 M16025 M12175 AF164486 U97669 AF058900 AF058881 AF058882 AF058883 AF058884 AF058885 AF058886 AF058887 AF058888 AF058889 AF058890 AF058891 AF058892 AF058893 AF058894 AF058895 AF058896 AF058897 AF058898 AF058899 AC004257 AC004663 L08692 X17530 M17421 X17533 BX005106 BX323836 CR456624 CU570696 AB011532 AL627123 BC031402 BC039980 BC058571 D63395 D86566 U95299 U89335 AL662884 BX284686 BX284927 CR812478 CR933878 AB023961 AB024520 AB024578 M80456 U43691 AF030001 CT009767 AB016771 AB016772 AB016773 AB016774 AB011539 AB231860 AL512413 AL513320 DQ227276 M33753 AE014297 AY118509 X05144 AF171092 BC058675 CR858597 AY358336 AK290029 AK295772 AK302477 AK304279 AK304721 AC093782 AC109361 CH471057 L38483 AF003837 U73936 AF028593 U61276 AK302554 AL035456 BC126205 BC126207 U77720 AB059656 AF397007 AF397008 AY035898 AY035899 AK050484 BC046642 BC068308
Proteomes
PRIDE
P35556
Q61555
P35555
Q61554
Q9TV36
Q75N90
+ More
P98133 Q14767 O35806 O08999 Q8CG19 Q28019 Q8N2S1 Q00918 Q14766 Q9NS15 Q61810 P21783 Q01705 G3I6Z6 Q07008 P46531 Q04721 O35516 Q9QW30 Q61982 P46530 P07207 Q9R172 Q9UM47 P10079 O88281 Q80V70 Q99466 P31695 O75095 Q2Q421 P10040 Q9QXX0 Q6UXI9 Q63722 P78504 Q91V88
P98133 Q14767 O35806 O08999 Q8CG19 Q28019 Q8N2S1 Q00918 Q14766 Q9NS15 Q61810 P21783 Q01705 G3I6Z6 Q07008 P46531 Q04721 O35516 Q9QW30 Q61982 P46530 P07207 Q9R172 Q9UM47 P10079 O88281 Q80V70 Q99466 P31695 O75095 Q2Q421 P10040 Q9QXX0 Q6UXI9 Q63722 P78504 Q91V88
Pfam
PF18193 Fibrillin_U_N
+ More
PF07645 EGF_CA
PF12661 hEGF
PF00683 TB
PF12662 cEGF
PF00008 EGF
PF12796 Ank_2
PF07684 NODP
PF00066 Notch
PF11936 DUF3454
PF06816 NOD
PF00023 Ank
PF01382 Avidin
PF00431 CUB
PF02210 Laminin_G_2
PF00053 Laminin_EGF
PF07974 EGF_2
PF01825 GPS
PF00002 7tm_2
PF00054 Laminin_G_1
PF01414 DSL
PF07657 MNNL
PF00629 MAM
PF07645 EGF_CA
PF12661 hEGF
PF00683 TB
PF12662 cEGF
PF00008 EGF
PF12796 Ank_2
PF07684 NODP
PF00066 Notch
PF11936 DUF3454
PF06816 NOD
PF00023 Ank
PF01382 Avidin
PF00431 CUB
PF02210 Laminin_G_2
PF00053 Laminin_EGF
PF07974 EGF_2
PF01825 GPS
PF00002 7tm_2
PF00054 Laminin_G_1
PF01414 DSL
PF07657 MNNL
PF00629 MAM
Interpro
IPR000152
EGF-type_Asp/Asn_hydroxyl_site
+ More
IPR017878 TB_dom
IPR013032 EGF-like_CS
IPR018097 EGF_Ca-bd_CS
IPR000742 EGF-like_dom
IPR009030 Growth_fac_rcpt_cys_sf
IPR026823 cEGF
IPR001881 EGF-like_Ca-bd_dom
IPR040872 Fibrillin_U_N
IPR036773 TB_dom_sf
IPR000800 Notch_dom
IPR024600 DUF3454_notch
IPR002110 Ankyrin_rpt
IPR036770 Ankyrin_rpt-contain_sf
IPR035993 Notch-like_dom_sf
IPR010660 Notch_NOD_dom
IPR022362 Notch_1
IPR020683 Ankyrin_rpt-contain_dom
IPR011656 Notch_NODP_dom
IPR008297 Notch
IPR022336 Notch_2
IPR022331 Notch_3
IPR036896 Avidin-like_sf
IPR005468 Avidin/str
IPR000859 CUB_dom
IPR005469 Avidin
IPR035914 Sperma_CUB_dom_sf
IPR017889 Avidin-like_CS
IPR001791 Laminin_G
IPR013320 ConA-like_dom_sf
IPR002049 Laminin_EGF
IPR011489 EMI_domain
IPR013111 EGF_extracell
IPR022355 Notch_4
IPR017983 GPCR_2_secretin-like_CS
IPR003056 GPCR_2_CD97
IPR017981 GPCR_2-like
IPR000832 GPCR_2_secretin-like
IPR000203 GPS
IPR001007 VWF_dom
IPR026219 Jagged/Serrate
IPR011651 Notch_ligand_N
IPR001774 DSL
IPR000998 MAM_dom
IPR017878 TB_dom
IPR013032 EGF-like_CS
IPR018097 EGF_Ca-bd_CS
IPR000742 EGF-like_dom
IPR009030 Growth_fac_rcpt_cys_sf
IPR026823 cEGF
IPR001881 EGF-like_Ca-bd_dom
IPR040872 Fibrillin_U_N
IPR036773 TB_dom_sf
IPR000800 Notch_dom
IPR024600 DUF3454_notch
IPR002110 Ankyrin_rpt
IPR036770 Ankyrin_rpt-contain_sf
IPR035993 Notch-like_dom_sf
IPR010660 Notch_NOD_dom
IPR022362 Notch_1
IPR020683 Ankyrin_rpt-contain_dom
IPR011656 Notch_NODP_dom
IPR008297 Notch
IPR022336 Notch_2
IPR022331 Notch_3
IPR036896 Avidin-like_sf
IPR005468 Avidin/str
IPR000859 CUB_dom
IPR005469 Avidin
IPR035914 Sperma_CUB_dom_sf
IPR017889 Avidin-like_CS
IPR001791 Laminin_G
IPR013320 ConA-like_dom_sf
IPR002049 Laminin_EGF
IPR011489 EMI_domain
IPR013111 EGF_extracell
IPR022355 Notch_4
IPR017983 GPCR_2_secretin-like_CS
IPR003056 GPCR_2_CD97
IPR017981 GPCR_2-like
IPR000832 GPCR_2_secretin-like
IPR000203 GPS
IPR001007 VWF_dom
IPR026219 Jagged/Serrate
IPR011651 Notch_ligand_N
IPR001774 DSL
IPR000998 MAM_dom
SUPFAM
Gene 3D
ProteinModelPortal
P35556.3
Q61555.2
P35555.4
Q61554.2
Q9TV36.1
Q75N90.3
+ More
P98133.2 Q14767.3 O35806.1 O08999.2 Q8CG19.2 Q28019.2 Q8N2S1.2 Q00918.1 Q14766.4 Q9NS15.4 Q61810.4 P21783.3 Q01705.3 G3I6Z6.2 A2RUV0.2 Q07008.3 P46531.4 Q04721.3 O35516.2 Q9QW30.1 Q61982.1 P46530.1 P07207.3 Q9R172.2 Q9UM47.2 P10079.2 B8JI71.2 O88281.1 Q80V70.3 Q99466.2 P31695.3 O75095.4 Q2Q421.2 P10040.3 Q9QXX0.1 Q5RBP1.1 Q6UXI9.3 Q63722.2 P78504.3 Q91V88.1
P98133.2 Q14767.3 O35806.1 O08999.2 Q8CG19.2 Q28019.2 Q8N2S1.2 Q00918.1 Q14766.4 Q9NS15.4 Q61810.4 P21783.3 Q01705.3 G3I6Z6.2 A2RUV0.2 Q07008.3 P46531.4 Q04721.3 O35516.2 Q9QW30.1 Q61982.1 P46530.1 P07207.3 Q9R172.2 Q9UM47.2 P10079.2 B8JI71.2 O88281.1 Q80V70.3 Q99466.2 P31695.3 O75095.4 Q2Q421.2 P10040.3 Q9QXX0.1 Q5RBP1.1 Q6UXI9.3 Q63722.2 P78504.3 Q91V88.1
PDB
5MS9
E-value=8.12981e-37,
Score=394
Ontologies
GO
GO:0030326
GO:0001527
GO:0060346
GO:0031012
GO:0035583
GO:0048048
GO:0030198
GO:0030501
GO:0043010
GO:0030023
GO:0062023
GO:0005509
GO:0045669
GO:0005576
GO:0005201
GO:0035108
GO:0005604
GO:0001656
GO:0008201
GO:0001501
GO:0034199
GO:0043687
GO:0006006
GO:0005179
GO:0035582
GO:0007507
GO:0005178
GO:0071560
GO:0005788
GO:0005615
GO:2001205
GO:0044877
GO:0042802
GO:0044267
GO:0010737
GO:0045671
GO:1990314
GO:0042593
GO:0048050
GO:0033627
GO:0001822
GO:0006605
GO:0097435
GO:0070062
GO:0019838
GO:0007179
GO:0050436
GO:0009306
GO:0005623
GO:0035904
GO:0048471
GO:0030425
GO:0038045
GO:0060976
GO:1901388
GO:0032991
GO:0005739
GO:0003281
GO:0005737
GO:0050431
GO:0032967
GO:0043025
GO:0071953
GO:0006457
GO:0045595
GO:0001558
GO:0030252
GO:0005024
GO:0007275
GO:0017015
GO:0005539
GO:0030162
GO:0071305
GO:0071374
GO:0036120
GO:0042060
GO:0071260
GO:1902462
GO:0036363
GO:2000741
GO:0060430
GO:0032331
GO:0045780
GO:0060349
GO:0046849
GO:0030502
GO:0050793
GO:0030154
GO:0005886
GO:0038023
GO:0000122
GO:0001525
GO:0060271
GO:0016021
GO:0005654
GO:0061314
GO:0021915
GO:0005794
GO:0035924
GO:0005112
GO:0003203
GO:0061384
GO:0005856
GO:0001889
GO:0043565
GO:0003162
GO:0010832
GO:0045668
GO:0045687
GO:0007050
GO:0000139
GO:0048708
GO:0030334
GO:0050434
GO:0045747
GO:0010628
GO:0060253
GO:1903849
GO:0045893
GO:0008593
GO:2000737
GO:2001027
GO:0055008
GO:0070374
GO:0045596
GO:0030514
GO:0003182
GO:0070986
GO:0072044
GO:0007492
GO:0048711
GO:0004888
GO:0003181
GO:0000978
GO:0001701
GO:0060317
GO:0045618
GO:0001228
GO:0003198
GO:0005789
GO:0048845
GO:0003209
GO:0045607
GO:0016324
GO:0009912
GO:0003344
GO:0048663
GO:0060843
GO:0005829
GO:0090051
GO:0031069
GO:0045967
GO:0045892
GO:0048103
GO:0060982
GO:0061344
GO:0010001
GO:0003252
GO:0001726
GO:0060548
GO:0001947
GO:0071372
GO:0008284
GO:0002052
GO:0032496
GO:0030182
GO:0002040
GO:0014031
GO:0003160
GO:0032495
GO:0021515
GO:0042127
GO:0072017
GO:0045608
GO:0030900
GO:0031410
GO:0035914
GO:0048873
GO:0003241
GO:0005634
GO:0007386
GO:0007221
GO:0003219
GO:0060528
GO:0060740
GO:0062043
GO:0008285
GO:0071944
GO:0002437
GO:0060956
GO:0045165
GO:0030216
GO:1901201
GO:0061419
GO:0008544
GO:0005887
GO:0030513
GO:0010629
GO:0003157
GO:0043086
GO:0014807
GO:0003332
GO:0035148
GO:0003222
GO:0050767
GO:0050679
GO:0060768
GO:0045665
GO:0005783
GO:0003180
GO:0043065
GO:0046579
GO:1902263
GO:0030027
GO:2000974
GO:0001837
GO:0043235
GO:0060411
GO:0046982
GO:0003256
GO:0035116
GO:0006959
GO:0060979
GO:0045955
GO:0046533
GO:0003151
GO:0045944
GO:0005912
GO:0060948
GO:0010718
GO:0003270
GO:0042246
GO:0072602
GO:0003208
GO:0031960
GO:0003192
GO:0007219
GO:0010614
GO:0003207
GO:0031100
GO:0003682
GO:0007409
GO:0003197
GO:2000811
GO:0070168
GO:0007283
GO:0030324
GO:0001708
GO:0007440
GO:0003700
GO:0090090
GO:0019899
GO:0003169
GO:0030335
GO:0045662
GO:0004857
GO:0010468
GO:0003213
GO:1902339
GO:0002051
GO:0046427
GO:0048754
GO:0060038
GO:0009986
GO:0001669
GO:0003184
GO:0060412
GO:0030279
GO:0010812
GO:0060842
GO:0050678
GO:0060045
GO:0003264
GO:0072144
GO:0002193
GO:0120163
GO:0045603
GO:0003214
GO:0007368
GO:0003273
GO:0045070
GO:0048709
GO:1901189
GO:0048715
GO:0031490
GO:0006357
GO:0050768
GO:0005796
GO:0006355
GO:0007420
GO:0097150
GO:0006367
GO:0006955
GO:0010838
GO:0006915
GO:0019827
GO:0001709
GO:0016020
GO:0060413
GO:0007399
GO:2001204
GO:0002315
GO:0030097
GO:0009887
GO:0043066
GO:2000249
GO:0072015
GO:0072104
GO:0005929
GO:0072014
GO:0035264
GO:0001890
GO:1990705
GO:0045672
GO:0035622
GO:0043011
GO:0060674
GO:0051059
GO:0072576
GO:0042742
GO:0061073
GO:0002011
GO:0072574
GO:0048844
GO:0015629
GO:0048661
GO:0021986
GO:0001756
GO:0021531
GO:0001840
GO:0048699
GO:0048259
GO:0031016
GO:0009952
GO:0048337
GO:0048936
GO:0031017
GO:0021514
GO:0042663
GO:0035907
GO:0021523
GO:0055016
GO:0002244
GO:0035204
GO:0048749
GO:1900087
GO:0008356
GO:0008347
GO:0043525
GO:0030720
GO:0060250
GO:0007519
GO:0030713
GO:0035003
GO:0022416
GO:0036099
GO:0007398
GO:0045466
GO:0007314
GO:0016360
GO:0050877
GO:0007473
GO:2000035
GO:0007298
GO:0005915
GO:2000048
GO:0005769
GO:0048665
GO:0007451
GO:0035153
GO:0061382
GO:0060571
GO:0007450
GO:0007015
GO:0035162
GO:0007346
GO:0005768
GO:0007422
GO:0048052
GO:0008045
GO:0035172
GO:0030139
GO:0007474
GO:0042691
GO:0048863
GO:0046666
GO:0035214
GO:0042689
GO:0008587
GO:0007405
GO:0046667
GO:0060289
GO:0046329
GO:0016348
GO:0042676
GO:0045316
GO:0035222
GO:0001745
GO:0035165
GO:0060288
GO:0048664
GO:0040008
GO:0035171
GO:0048190
GO:0007498
GO:0007464
GO:0036011
GO:0014019
GO:0008407
GO:0042688
GO:0048803
GO:1902692
GO:1990433
GO:0030708
GO:0007297
GO:0002213
GO:0007476
GO:0048542
GO:0046843
GO:0007446
GO:0007400
GO:0009608
GO:0016333
GO:0016330
GO:0007419
GO:0043697
GO:0035167
GO:0007423
GO:0007411
GO:0030718
GO:0007616
GO:0007455
GO:0006110
GO:0046331
GO:0008340
GO:0007403
GO:0009950
GO:0046716
GO:0042067
GO:0036335
GO:0051489
GO:0007391
GO:0007447
GO:0005770
GO:0030707
GO:0042686
GO:0048477
GO:0007478
GO:0061331
GO:0007521
GO:0007293
GO:0045296
GO:0045746
GO:0032579
GO:0033165
GO:0050896
GO:0007601
GO:0046549
GO:0035845
GO:0045494
GO:0045602
GO:0001886
GO:0001944
GO:0001569
GO:0035278
GO:0001763
GO:0030879
GO:0070613
GO:0045446
GO:0045766
GO:0032880
GO:0032587
GO:0006954
GO:0007166
GO:0031256
GO:0071621
GO:0016477
GO:0004930
GO:0007155
GO:0035374
GO:0007189
GO:0035332
GO:0016332
GO:0003383
GO:0061024
GO:0005080
GO:0046621
GO:0007163
GO:0106036
GO:0035239
GO:0061336
GO:0007443
GO:0007435
GO:0005819
GO:0046664
GO:0007424
GO:0032435
GO:0035090
GO:0045570
GO:0098813
GO:0016028
GO:0033157
GO:0061541
GO:0016334
GO:0051642
GO:0046665
GO:0034332
GO:0042051
GO:0045186
GO:0030507
GO:0035088
GO:0045176
GO:0050821
GO:0045218
GO:0060438
GO:0051301
GO:0042052
GO:0016327
GO:0035002
GO:0061309
GO:0072006
GO:0003215
GO:0001974
GO:0045177
GO:0022408
GO:0042491
GO:0002456
GO:0072070
GO:0035909
GO:0061156
GO:0001953
GO:0045599
GO:0048018
GO:0061444
GO:0048839
GO:0005543
GO:0045639
GO:0030336
GO:0097195
GO:0001657
GO:2000721
GO:0034678
GO:0001658
GO:0010811
GO:0010694
GO:0045987
GO:0007160
GO:0045184
GO:0030485
GO:0071356
GO:0033631
GO:0030511
GO:0008083
GO:0005198
GO:0045445
GO:0005515
GO:0020037
GO:0005524
GO:0003909
GO:0051103
GO:0000049
GO:0005975
GO:0004553
Topology
Subcellular location
Secreted Fibrillin-1 and Asprosin chains are still linked together during the secretion from cells, but are subsequently separated by furin (PubMed:24982166). With evidence from 68 publications.
Extracellular space
Extracellular matrix
Cell membrane
Nucleus Following proteolytical processing NICD is translocated to the nucleus. With evidence from 4 publications.
Cytoplasm Following proteolytical processing NICD is translocated to the nucleus. Retained at the cytoplasm by TCIM (PubMed:25985737). With evidence from 26 publications.
Endosome
Cytoplasmic vesicle
Hyaline layer
Apical lamina
Cell projection Localizes to discrete puncta at, or adjacent to, the photoreceptor connecting cilium (PubMed:27737822, PubMed:30052645). May localize to the cilium axoneme (By similarity). May also be expressed in the interphotoreceptor extracellular matrix (By similarity). Unlike the primate ortholog, does not appear to be expressed in the photoreceptor outer segment (PubMed:27737822). With evidence from 1 publications.
Cilium Localizes to discrete puncta at, or adjacent to, the photoreceptor connecting cilium (PubMed:27737822, PubMed:30052645). May localize to the cilium axoneme (By similarity). May also be expressed in the interphotoreceptor extracellular matrix (By similarity). Unlike the primate ortholog, does not appear to be expressed in the photoreceptor outer segment (PubMed:27737822). With evidence from 1 publications.
Cytoskeleton Localizes to discrete puncta at, or adjacent to, the photoreceptor connecting cilium (PubMed:27737822, PubMed:30052645). May localize to the cilium axoneme (By similarity). May also be expressed in the interphotoreceptor extracellular matrix (By similarity). Unlike the primate ortholog, does not appear to be expressed in the photoreceptor outer segment (PubMed:27737822). With evidence from 1 publications.
Cilium axoneme Localizes to discrete puncta at, or adjacent to, the photoreceptor connecting cilium (PubMed:27737822, PubMed:30052645). May localize to the cilium axoneme (By similarity). May also be expressed in the interphotoreceptor extracellular matrix (By similarity). Unlike the primate ortholog, does not appear to be expressed in the photoreceptor outer segment (PubMed:27737822). With evidence from 1 publications.
Interphotoreceptor matrix Localizes to discrete puncta at, or adjacent to, the photoreceptor connecting cilium (PubMed:27737822, PubMed:30052645). May localize to the cilium axoneme (By similarity). May also be expressed in the interphotoreceptor extracellular matrix (By similarity). Unlike the primate ortholog, does not appear to be expressed in the photoreceptor outer segment (PubMed:27737822). With evidence from 1 publications.
Ruffle membrane Localized at the leading edge of migrating cells. With evidence from 1 publications.
Apical cell membrane Specifically localized to the apical membrane (PubMed:11076972, PubMed:2344615). Expressed in a mesh punctate pattern that overlaps with metaphase spindle microtubules (PubMed:25065591). In tracheal cells, colocalizes with apn in the subapical region, a small region of the apical membrane apical to the adherens junctions (PubMed:30645584). With evidence from 7 publications.
Spindle Specifically localized to the apical membrane (PubMed:11076972, PubMed:2344615). Expressed in a mesh punctate pattern that overlaps with metaphase spindle microtubules (PubMed:25065591). In tracheal cells, colocalizes with apn in the subapical region, a small region of the apical membrane apical to the adherens junctions (PubMed:30645584). With evidence from 7 publications.
Membrane
SignalP
Position: 1 - 18,
Likelihood: 0.992465
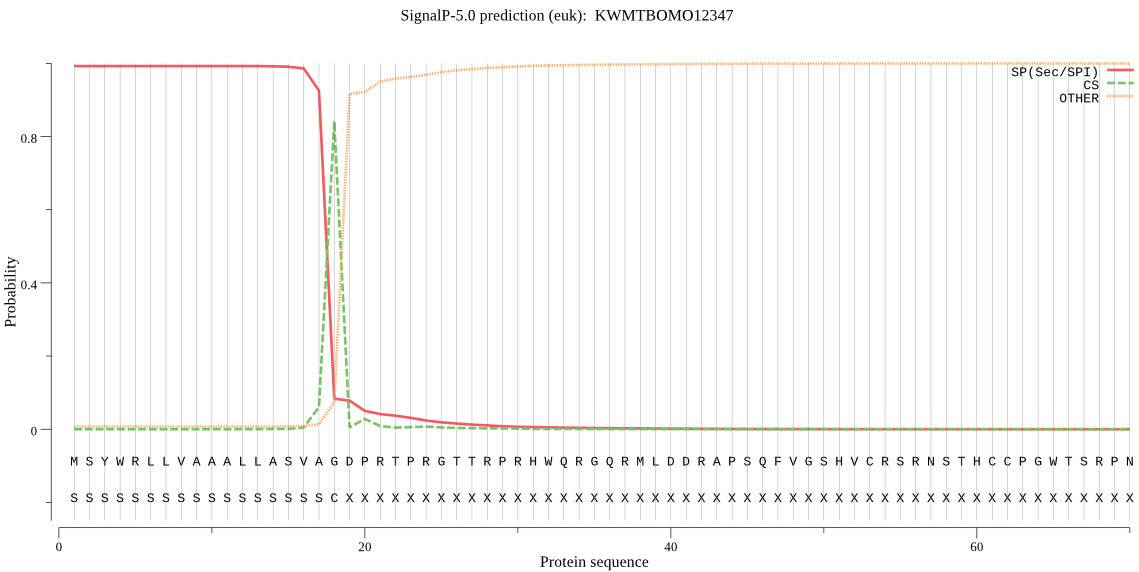
Length:
3016
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0684500000000002
Exp number, first 60 AAs:
0.06763
Total prob of N-in:
0.00376
outside
1 - 3016
Population Genetic Test Statistics
Pi
2.163756
Theta
162.80826
Tajima's D
0.643411
CLR
0.46204
CSRT
0.555822208889555
Interpretation
Uncertain
