Gene
KWMTBOMO12342
Pre Gene Modal
BGIBMGA012385
Annotation
cytochrome_P450_6AB4_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.2 Mitochondrial Reliability : 1.396 PlasmaMembrane Reliability : 1.613
Sequence
CDS
ATGCTCACCGCCGCGATATTCGTTATCATTGTCGCTCTTGTTTATTTGTACTCAACTAGAACGTTCAGATACTGGGAGAAAAGGGGCATCAAACATGATAAGCCTGTACCCTTCTTCGGCACCGATTCAGAGGGATATCTACTCAGAAAAAGCATGACCCAAACTGCTGTGGATGCATATCTCAAATACCCCAATGAAAAAGTCATCGGTTTCTTCAGATCCTCAAGGCCTGAACTAATCATCAGAGACCCGGATATTATAAAACGAATTCTAACAACAGACTTCGCGTATTTCTATCCAAGGGGACTGAATCCACATAAAAAGGTAATTGAGCCGTTGATGCGTAACCTGTTTTTCGCTGATGGGGATCTGTGGCGTCTTCTACGTCAACGTATGACACCTGCATTTACCAGCGGGAAATTGCGTGCCATGTTTCCTCTGGTCGTGGAAAGAGCTGAAAGACTTCAATCTCGGACCCTTGAGATTGCTTCACAACCTCTAGATGCTAGAGAACTAATGGCACGTTACACGACAGATTTCATAGGCGCTTGCGGCTTTGGCCTCGATGCAGACTCACTGAACGACGAAGATTCAGCATTCAGAAAGCTTGGAGCGGCCATTTTCAACATAACTGTTCAGCAAGCAATCGTGGCTGCCCTCAAAGAGATATTCCCTGGGATATTCAAGCATTTTAAGTACTCATCGAAAATGAAAGGAGAAATCGTAGTGGAATCTATTGAGAAAATGAAACCAGATGGCACTTCTGAAGTCGTTAGAATGGAGTTAACAGAACAACTGCTAGCAGCTCAAGTATTCATCTTCTTTGCAGCTGGCTTTGAAACTTCGTCGTCTGCCACTAGCTTCACGCTCCATCAACTTGCGTTCCATCCTGAGATCCAAGAGAAAGTGCAAAAAGAACTAGATCAGGTTCTCGCCAAATATAACAACAAGCTGTGCTACGATGCGATCAAAGAGATGAGATACTTGGAGAGTGCGTTTAAAGAGGCCATGAGAATGTTTCCGTCGCTCGGTTTCTTGATTAGGGAGTGCGCTCGTCAATACACATTCCCGGAGTTAAACTTGACTATAGACGAAGGCGTAGGTATAATCATACCTCTCCAGGCCTTGCACAATGACCCTGAATATTTTGATAGTCCGAATGAATTCCGACCAGAAAGATTCATGCCCTCTGAATATAATCATAACAAAACGAAATTTGTCTACTTACCTTTTGGCGATGGACCACGTGGTTGCATCGGTGCTCGCCTTGGCTTAATGCAGTCCTTGGCAGGCTTGGCAGCCGTCCTGTCCAAGTTCACGGTGAAGCCAGCTCCGAACACAAAGCGTCATCCGGTCGTTGAACCCAAATCCAGCATCGTGCAGAGCTTCAAAGATGACCTGCCTCTGCTCTTCATTGAGAGGACAAAGCCCTGA
Protein
MLTAAIFVIIVALVYLYSTRTFRYWEKRGIKHDKPVPFFGTDSEGYLLRKSMTQTAVDAYLKYPNEKVIGFFRSSRPELIIRDPDIIKRILTTDFAYFYPRGLNPHKKVIEPLMRNLFFADGDLWRLLRQRMTPAFTSGKLRAMFPLVVERAERLQSRTLEIASQPLDARELMARYTTDFIGACGFGLDADSLNDEDSAFRKLGAAIFNITVQQAIVAALKEIFPGIFKHFKYSSKMKGEIVVESIEKMKPDGTSEVVRMELTEQLLAAQVFIFFAAGFETSSSATSFTLHQLAFHPEIQEKVQKELDQVLAKYNNKLCYDAIKEMRYLESAFKEAMRMFPSLGFLIRECARQYTFPELNLTIDEGVGIIIPLQALHNDPEYFDSPNEFRPERFMPSEYNHNKTKFVYLPFGDGPRGCIGARLGLMQSLAGLAAVLSKFTVKPAPNTKRHPVVEPKSSIVQSFKDDLPLLFIERTKP
Summary
Cofactor
heme
Similarity
Belongs to the cytochrome P450 family.
Uniprot
L0N4S4
A0MNW5
D0FZ09
A3RIC3
H9JS75
A0FGR5
+ More
L0N4I6 L0N5F4 J7FJ24 A0A1L5BWN2 A0A248QFZ8 G1CQY9 A0A0L7KN22 A0A2A4JQP9 A0A0L7KNZ8 A0A194RAH0 A0A0K8TUI9 A0A1E1WF52 H9JS76 A0A1E1WEJ2 A0A0K8TVR7 A0A2W1BFR2 X5D4F8 A0A2W1BEP8 A0A1E1W8J2 A0A1E1W3A5 A0A068EWP6 A0A248QEH3 A0A2A4J2N5 A0A194Q0W4 G8GPR2 A0A2H1VLH9 A0A194PDG1 A0A0N0PCI0 A0A1V0D9D2 A0A3S2LXS3 J7FJY1 A0A1E1WKY6 A0A1E1W3D4 A0A0N1I9K3 A0A2H1VY40 A0A194PCU6 A0A0L7KIV2 X5DB34 A0A3Q9D0K0 X5DAY3 S4VDQ0 A0A194PIY6 A0A194RIE1 A0A1E1WBG3 A0A0N0PFJ3 A0A3S2PHF3 A0A248QHD5 X5D9B1 A0A194PED0 A0A0L7KNL0 A0A0N1IJR7 A0A3Q8EMJ5 A0A2A4JPX0 A0A2H1WVY7 A0A212EYB4 A0A0L7KIX6 A0A2W1BTE1 X5DSZ6 A0A0N0PEX0 A0A0K8TUJ3 A0A286MXM3 A0A3S2LXP2 A0A0K8TUM3 X5DT00 A0A0A7DH67 X5DAY7 A0A0A7DH60 A0A2H1VKD6 A0A248QEX2 A0A0K8TV73 A0A1E1WQ45 L7R5T3 D5L0M9 A1E5T6 A0A0L7L1B3 A1E5T7 Q7YZS3 L7R4I9 H9JRC9 W5VXT0 A0A0K8TUJ7 A0A3G1T1F1 A0A2H1WPX9 J7FIS9 A0A0L7L0W5 A0A1E1WTK8 A0A248QJ02 S4VGG2 A0A2H1X2Q0 A0A0L7KP53 A0A346IHY3 A0A1V0D9D5 X5DQP4 A0A0L7KV53
L0N4I6 L0N5F4 J7FJ24 A0A1L5BWN2 A0A248QFZ8 G1CQY9 A0A0L7KN22 A0A2A4JQP9 A0A0L7KNZ8 A0A194RAH0 A0A0K8TUI9 A0A1E1WF52 H9JS76 A0A1E1WEJ2 A0A0K8TVR7 A0A2W1BFR2 X5D4F8 A0A2W1BEP8 A0A1E1W8J2 A0A1E1W3A5 A0A068EWP6 A0A248QEH3 A0A2A4J2N5 A0A194Q0W4 G8GPR2 A0A2H1VLH9 A0A194PDG1 A0A0N0PCI0 A0A1V0D9D2 A0A3S2LXS3 J7FJY1 A0A1E1WKY6 A0A1E1W3D4 A0A0N1I9K3 A0A2H1VY40 A0A194PCU6 A0A0L7KIV2 X5DB34 A0A3Q9D0K0 X5DAY3 S4VDQ0 A0A194PIY6 A0A194RIE1 A0A1E1WBG3 A0A0N0PFJ3 A0A3S2PHF3 A0A248QHD5 X5D9B1 A0A194PED0 A0A0L7KNL0 A0A0N1IJR7 A0A3Q8EMJ5 A0A2A4JPX0 A0A2H1WVY7 A0A212EYB4 A0A0L7KIX6 A0A2W1BTE1 X5DSZ6 A0A0N0PEX0 A0A0K8TUJ3 A0A286MXM3 A0A3S2LXP2 A0A0K8TUM3 X5DT00 A0A0A7DH67 X5DAY7 A0A0A7DH60 A0A2H1VKD6 A0A248QEX2 A0A0K8TV73 A0A1E1WQ45 L7R5T3 D5L0M9 A1E5T6 A0A0L7L1B3 A1E5T7 Q7YZS3 L7R4I9 H9JRC9 W5VXT0 A0A0K8TUJ7 A0A3G1T1F1 A0A2H1WPX9 J7FIS9 A0A0L7L0W5 A0A1E1WTK8 A0A248QJ02 S4VGG2 A0A2H1X2Q0 A0A0L7KP53 A0A346IHY3 A0A1V0D9D5 X5DQP4 A0A0L7KV53
Pubmed
EMBL
AK343203
BAM73889.1
EF017798
ABK27873.1
AB498806
BAI47531.1
+ More
EF415299 ABN71370.1 BABH01033060 DQ993198 ABJ97708.1 AK289287 BAM73814.1 AK289286 BAM73813.1 JX310081 AFP20592.1 KX827419 APL97605.1 KX443423 ASO97998.1 JF829002 AEL87782.1 JTDY01008220 KOB64673.1 NWSH01000809 PCG74096.1 KOB64674.1 KQ460473 KPJ14499.1 GCVX01000071 JAI18159.1 GDQN01005420 JAT85634.1 BABH01033062 GDQN01005707 GDQN01002158 JAT85347.1 JAT88896.1 GCVX01000070 JAI18160.1 KZ150094 PZC73628.1 KF701131 AHW57301.1 PZC73629.1 GDQN01007873 GDQN01001117 JAT83181.1 JAT89937.1 GDQN01011078 GDQN01009598 JAT79976.1 JAT81456.1 KM016734 AID54886.1 KX443424 ASO97999.1 NWSH01003644 PCG65996.1 KQ459591 KPI97045.1 JN176570 AET11927.1 ODYU01003027 SOQ41282.1 KQ459606 KPI91048.1 KQ460545 KPJ14074.1 KY212041 ARA91605.1 RSAL01000123 RVE46645.1 JX310080 AFP20591.1 GDQN01005674 GDQN01003533 JAT85380.1 JAT87521.1 GDQN01009559 JAT81495.1 KPJ14075.1 ODYU01004989 SOQ45412.1 KPI91047.1 JTDY01009584 KOB63273.1 KF701190 AHW57360.1 MH644187 AZP54635.1 KF701130 AHW57300.1 KC789747 AGO62002.1 KPI91050.1 KQ460205 KPJ17095.1 GDQN01006843 JAT84211.1 LADJ01036943 KPJ21304.1 RSAL01000032 RVE51553.1 KX443422 ASO97997.1 KF701132 AHW57302.1 KPI91049.1 KOB64675.1 KPJ21305.1 MG697223 AVR93642.1 PCG74097.1 ODYU01011431 SOQ57167.1 AGBW02011568 OWR46482.1 KOB63272.1 KZ149941 PZC76905.1 KF701129 AHW57299.1 KQ459620 KPJ20541.1 GCVX01000066 JAI18164.1 MF684341 ASX93975.1 RSAL01000131 RVE46388.1 GCVX01000048 JAI18182.1 KF701134 AHW57304.1 KJ645915 AIJ00772.1 KF701135 AHW57305.1 KJ645916 AIJ00773.1 ODYU01003028 SOQ41283.1 KX443425 ASO98000.1 GCVX01000047 JAI18183.1 GDQN01011046 GDQN01002027 JAT80008.1 JAT89027.1 JQ901385 AGB93713.1 GU731534 ADE05584.1 EF111023 ABL60877.1 JTDY01003740 KOB69064.1 EF111024 ABL60878.1 AY295773 AAP83688.1 JQ901386 AGB93714.1 BABH01016132 BABH01016133 KF853195 AHH92933.1 GCVX01000061 JAI18169.1 MG846941 AXY94793.1 ODYU01010161 SOQ55088.1 JX310074 AFP20585.1 JTDY01003692 KOB69138.1 GDQN01000714 JAT90340.1 KX443426 ASO98001.1 KC789748 AGO62003.1 ODYU01012980 SOQ59557.1 JTDY01007935 KOB64846.1 MH138197 AXP17138.1 KY212046 ARA91610.1 KF701133 AHW57303.1 JTDY01005488 KOB66951.1
EF415299 ABN71370.1 BABH01033060 DQ993198 ABJ97708.1 AK289287 BAM73814.1 AK289286 BAM73813.1 JX310081 AFP20592.1 KX827419 APL97605.1 KX443423 ASO97998.1 JF829002 AEL87782.1 JTDY01008220 KOB64673.1 NWSH01000809 PCG74096.1 KOB64674.1 KQ460473 KPJ14499.1 GCVX01000071 JAI18159.1 GDQN01005420 JAT85634.1 BABH01033062 GDQN01005707 GDQN01002158 JAT85347.1 JAT88896.1 GCVX01000070 JAI18160.1 KZ150094 PZC73628.1 KF701131 AHW57301.1 PZC73629.1 GDQN01007873 GDQN01001117 JAT83181.1 JAT89937.1 GDQN01011078 GDQN01009598 JAT79976.1 JAT81456.1 KM016734 AID54886.1 KX443424 ASO97999.1 NWSH01003644 PCG65996.1 KQ459591 KPI97045.1 JN176570 AET11927.1 ODYU01003027 SOQ41282.1 KQ459606 KPI91048.1 KQ460545 KPJ14074.1 KY212041 ARA91605.1 RSAL01000123 RVE46645.1 JX310080 AFP20591.1 GDQN01005674 GDQN01003533 JAT85380.1 JAT87521.1 GDQN01009559 JAT81495.1 KPJ14075.1 ODYU01004989 SOQ45412.1 KPI91047.1 JTDY01009584 KOB63273.1 KF701190 AHW57360.1 MH644187 AZP54635.1 KF701130 AHW57300.1 KC789747 AGO62002.1 KPI91050.1 KQ460205 KPJ17095.1 GDQN01006843 JAT84211.1 LADJ01036943 KPJ21304.1 RSAL01000032 RVE51553.1 KX443422 ASO97997.1 KF701132 AHW57302.1 KPI91049.1 KOB64675.1 KPJ21305.1 MG697223 AVR93642.1 PCG74097.1 ODYU01011431 SOQ57167.1 AGBW02011568 OWR46482.1 KOB63272.1 KZ149941 PZC76905.1 KF701129 AHW57299.1 KQ459620 KPJ20541.1 GCVX01000066 JAI18164.1 MF684341 ASX93975.1 RSAL01000131 RVE46388.1 GCVX01000048 JAI18182.1 KF701134 AHW57304.1 KJ645915 AIJ00772.1 KF701135 AHW57305.1 KJ645916 AIJ00773.1 ODYU01003028 SOQ41283.1 KX443425 ASO98000.1 GCVX01000047 JAI18183.1 GDQN01011046 GDQN01002027 JAT80008.1 JAT89027.1 JQ901385 AGB93713.1 GU731534 ADE05584.1 EF111023 ABL60877.1 JTDY01003740 KOB69064.1 EF111024 ABL60878.1 AY295773 AAP83688.1 JQ901386 AGB93714.1 BABH01016132 BABH01016133 KF853195 AHH92933.1 GCVX01000061 JAI18169.1 MG846941 AXY94793.1 ODYU01010161 SOQ55088.1 JX310074 AFP20585.1 JTDY01003692 KOB69138.1 GDQN01000714 JAT90340.1 KX443426 ASO98001.1 KC789748 AGO62003.1 ODYU01012980 SOQ59557.1 JTDY01007935 KOB64846.1 MH138197 AXP17138.1 KY212046 ARA91610.1 KF701133 AHW57303.1 JTDY01005488 KOB66951.1
Proteomes
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
L0N4S4
A0MNW5
D0FZ09
A3RIC3
H9JS75
A0FGR5
+ More
L0N4I6 L0N5F4 J7FJ24 A0A1L5BWN2 A0A248QFZ8 G1CQY9 A0A0L7KN22 A0A2A4JQP9 A0A0L7KNZ8 A0A194RAH0 A0A0K8TUI9 A0A1E1WF52 H9JS76 A0A1E1WEJ2 A0A0K8TVR7 A0A2W1BFR2 X5D4F8 A0A2W1BEP8 A0A1E1W8J2 A0A1E1W3A5 A0A068EWP6 A0A248QEH3 A0A2A4J2N5 A0A194Q0W4 G8GPR2 A0A2H1VLH9 A0A194PDG1 A0A0N0PCI0 A0A1V0D9D2 A0A3S2LXS3 J7FJY1 A0A1E1WKY6 A0A1E1W3D4 A0A0N1I9K3 A0A2H1VY40 A0A194PCU6 A0A0L7KIV2 X5DB34 A0A3Q9D0K0 X5DAY3 S4VDQ0 A0A194PIY6 A0A194RIE1 A0A1E1WBG3 A0A0N0PFJ3 A0A3S2PHF3 A0A248QHD5 X5D9B1 A0A194PED0 A0A0L7KNL0 A0A0N1IJR7 A0A3Q8EMJ5 A0A2A4JPX0 A0A2H1WVY7 A0A212EYB4 A0A0L7KIX6 A0A2W1BTE1 X5DSZ6 A0A0N0PEX0 A0A0K8TUJ3 A0A286MXM3 A0A3S2LXP2 A0A0K8TUM3 X5DT00 A0A0A7DH67 X5DAY7 A0A0A7DH60 A0A2H1VKD6 A0A248QEX2 A0A0K8TV73 A0A1E1WQ45 L7R5T3 D5L0M9 A1E5T6 A0A0L7L1B3 A1E5T7 Q7YZS3 L7R4I9 H9JRC9 W5VXT0 A0A0K8TUJ7 A0A3G1T1F1 A0A2H1WPX9 J7FIS9 A0A0L7L0W5 A0A1E1WTK8 A0A248QJ02 S4VGG2 A0A2H1X2Q0 A0A0L7KP53 A0A346IHY3 A0A1V0D9D5 X5DQP4 A0A0L7KV53
L0N4I6 L0N5F4 J7FJ24 A0A1L5BWN2 A0A248QFZ8 G1CQY9 A0A0L7KN22 A0A2A4JQP9 A0A0L7KNZ8 A0A194RAH0 A0A0K8TUI9 A0A1E1WF52 H9JS76 A0A1E1WEJ2 A0A0K8TVR7 A0A2W1BFR2 X5D4F8 A0A2W1BEP8 A0A1E1W8J2 A0A1E1W3A5 A0A068EWP6 A0A248QEH3 A0A2A4J2N5 A0A194Q0W4 G8GPR2 A0A2H1VLH9 A0A194PDG1 A0A0N0PCI0 A0A1V0D9D2 A0A3S2LXS3 J7FJY1 A0A1E1WKY6 A0A1E1W3D4 A0A0N1I9K3 A0A2H1VY40 A0A194PCU6 A0A0L7KIV2 X5DB34 A0A3Q9D0K0 X5DAY3 S4VDQ0 A0A194PIY6 A0A194RIE1 A0A1E1WBG3 A0A0N0PFJ3 A0A3S2PHF3 A0A248QHD5 X5D9B1 A0A194PED0 A0A0L7KNL0 A0A0N1IJR7 A0A3Q8EMJ5 A0A2A4JPX0 A0A2H1WVY7 A0A212EYB4 A0A0L7KIX6 A0A2W1BTE1 X5DSZ6 A0A0N0PEX0 A0A0K8TUJ3 A0A286MXM3 A0A3S2LXP2 A0A0K8TUM3 X5DT00 A0A0A7DH67 X5DAY7 A0A0A7DH60 A0A2H1VKD6 A0A248QEX2 A0A0K8TV73 A0A1E1WQ45 L7R5T3 D5L0M9 A1E5T6 A0A0L7L1B3 A1E5T7 Q7YZS3 L7R4I9 H9JRC9 W5VXT0 A0A0K8TUJ7 A0A3G1T1F1 A0A2H1WPX9 J7FIS9 A0A0L7L0W5 A0A1E1WTK8 A0A248QJ02 S4VGG2 A0A2H1X2Q0 A0A0L7KP53 A0A346IHY3 A0A1V0D9D5 X5DQP4 A0A0L7KV53
PDB
6MJM
E-value=9.86319e-53,
Score=524
Ontologies
GO
Topology
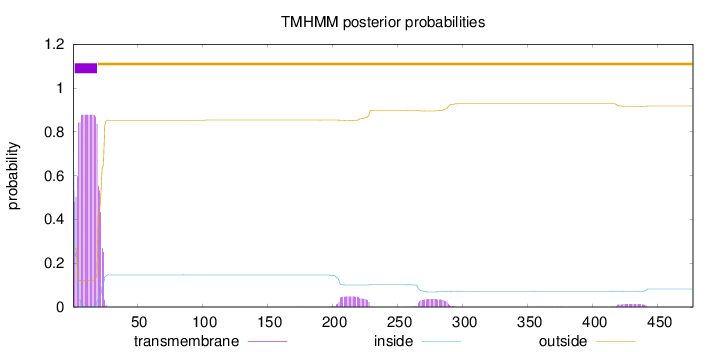
Length:
477
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.83145
Exp number, first 60 AAs:
16.70498
Total prob of N-in:
0.73192
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 19
outside
20 - 477
Population Genetic Test Statistics
Pi
211.884598
Theta
175.045657
Tajima's D
0.718034
CLR
0
CSRT
0.572271386430678
Interpretation
Uncertain
