Pre Gene Modal
BGIBMGA009181
Annotation
PREDICTED:_DNA_ligase_1_[Amyelois_transitella]
Full name
DNA ligase
+ More
DNA ligase 1
DNA ligase 1
Alternative Name
DNA ligase I
Polydeoxyribonucleotide synthase [ATP] 1
Polydeoxyribonucleotide synthase [ATP] 1
Location in the cell
Nuclear Reliability : 2.928
Sequence
CDS
ATGGCTCAAAGGAGTATCACGTCATTCTTCAAGAAAACTCCATTTACACCTATGGAAATAAAAAGTAATGGTGGATCCAAAGTAAGCCATGTAAATGGGGACACATCACCCATATCCGACGAAAAAGACAAGCCTTCTGAAAGACAACAATCCAACAGCATAGATAATGACGGTGACTCCCAAGATTCTTTGAAACCGAAAAATAAAGCTAAACGACAACGTGTGATAAGCTCTAGTAGTGAATCAGAATCTCCAGTTAAAGTGAAAAAGGAAGACCTTTCAGACAATGATGACTCTATAACTTTAAACAAGAGCGTAAACAAAACTTATGACTCACCAAAAAATAAAAAAGCTAAAATAAAAAAAGAAATCAAAAGCCCATCACCTAAAAAGGGTGCAAAGAGTTTGTCTCGTGATGAGGTAAAAGAAAACAGCTTAAAATTGAAGATTGTTGGATCAAAGGAAAAAGACAATATTGTGAAAAATACCGAGCATATTGATAATATTGAAAATGAGAGTGAAGAAATACAGAAAGTAGCAGAAAATGCATACAATCCATCAAAATCAAAATACCACCCTATAAAAGATGCTTGCTGGTCTAAAGGCGACCCAGTGCCGTATCTTGCATTAGCTAAGACTTTGGATCTTATAGAAGGAACATCAGCTCGACTGAAGATAATAGAAATATTAAGTAATTTTTATAGGTCTGTAATGGTGTTGACTTCAGAAGACTTGTTGCCTTGTATTTACTTGTGCTTAAATCAACTAGCACCAGCATACCACAGTTTGGAGCTTGGCATAGCAGAAACGTACCTAATGAAAGCCATAGGCCAGTGTACAGGCCGTACCCTAGCACAAGTGAAGTCTGCAGTACAGAAGTGTGGGGATATGGGAAGAGTGGCAGAACAGGCTCGAGCCGCACAAAAGACAATGTTTGCGGCCCCTAAACTTCAGATAAGAAAGGTTTTTGCTGCACTAAAAGAAATTGCTCACATGACTGGTCAAGCATCAGTGAATAAGAAAATAAATAAAATCCAGTCCTTGTATGTCGCCTGCAGAGATTGTGAAGCACGATTCCTAATAAGGTCCCTGGAGGGGAAGCTGCGCATCGGCCTGGCGGAGCAGTCGGTGCTGCACGCGCTGGCGCTGGCCGTCGCCACCACGCCGCCCGGACCCAACCTGCAGCTCGACGCGGCGCAGCACATGTCCGCCGACAAGTTCAAGGCGCTGGTGGAGCAGCACGTGGCGGTGGTGCGGCGCACGTACTGCGAGTGTCCCGACTGGGCCGAGCTGCTGCACGAGCTGCTGCGCGGCGGCGTGGCGGCGCTGGCCTCCCGCTGCAGCATGCGCGTCGGCGTGCCGCTGCGCCCCATGCTCGCGCACCCCACCCGCGCCGTGCGCGACGTGCTCGCCAGGTTTGAGAATGAAGAGTTCACCTGCGAGTACAAGTACGACGGCGAGCGTGCCCAGATCCACGTGCCCGCCGCGCCCGACGACAAGCCGGACTTCGCCAACGCTGCCATATTCAGCAGGAACCAAGAAAATAATACCGCCAAGTACCCGGACGTGCTGAGTCGCCTGCCCACGCTGCTCAAGGAGGGCGTGTCCAGCTGCGTGCTGGACTGCGAGGCCGTCGCCTTCGACGCCGCCAACGGCAAGCTGCTGCCCTTCCAGGTGCTGTCGAGCCGCAAGCGTGCGTGCGCGAGTGCGGAGGACGTGCGCGTGCCGGTCTGCGTGTTCGTGTTCGACCTGCTGTACCTCAACGGCGCCCCGCTGGTGGGCAGCACGCTGGCGCACCGCCGCAAGCTGCTGCGGGAACACTTCCGCACCCAGCCCGGTACATGGCAGTTCGCCACGTCCAAAGACTGCAGCACTATCGAGGAAGTGCAGCTGTTCCTTGAAGAAGCTGTGAAAGAAAGCTGCGAGGGACTCATGGTGAAGATGCTCACTGGTGACAATTCGAAATATGATATAGCTAGGAGGTCACACAATTGGCTAAAGTTGAAGAAAGACTACTTGGAAGGCGTCGGAGACACGGTGGACGCTGTCATTATCGGAGCCTACCGCGGCAAGGGCAAGCGCGCCGGCGTCTACGGAGGGTTCCTGGTCGCGTGCTACGACCCCGAGTCCGAGCAGTTCCAGTCGCTGTGCAAGCTGGGGACCGGCTTCTCCGATGAGGACCTGCAGGCCTTGTCCGACATGCTGAAGGAACACATTATTGATTCTCCGAGAAACTACTACAACTACGACAGCTCCCACTCCGCCGACGTATGGACCAGCGCCGCCGTGGTGGTGGAGGTGCGCTGCGCAGACCTGTCCCTCTCGCCCGCGCACCGCGCCGCCATCGCGCGCCTGCACCCGCACAAGGGCGTGTCACTCCGCTTCCCCAGGTTCGTGCGGGTGCGGGAGGACAAGGCGGCGGAGCAGGCCACCACGGCGGCGCAGGTGGCGCAGCTCTACCTGGCGCAGGACCAGATCAAGAACCAAGTCAAACAACCTGACGCCGCTCACGAAGACGACTTCTACTGA
Protein
MAQRSITSFFKKTPFTPMEIKSNGGSKVSHVNGDTSPISDEKDKPSERQQSNSIDNDGDSQDSLKPKNKAKRQRVISSSSESESPVKVKKEDLSDNDDSITLNKSVNKTYDSPKNKKAKIKKEIKSPSPKKGAKSLSRDEVKENSLKLKIVGSKEKDNIVKNTEHIDNIENESEEIQKVAENAYNPSKSKYHPIKDACWSKGDPVPYLALAKTLDLIEGTSARLKIIEILSNFYRSVMVLTSEDLLPCIYLCLNQLAPAYHSLELGIAETYLMKAIGQCTGRTLAQVKSAVQKCGDMGRVAEQARAAQKTMFAAPKLQIRKVFAALKEIAHMTGQASVNKKINKIQSLYVACRDCEARFLIRSLEGKLRIGLAEQSVLHALALAVATTPPGPNLQLDAAQHMSADKFKALVEQHVAVVRRTYCECPDWAELLHELLRGGVAALASRCSMRVGVPLRPMLAHPTRAVRDVLARFENEEFTCEYKYDGERAQIHVPAAPDDKPDFANAAIFSRNQENNTAKYPDVLSRLPTLLKEGVSSCVLDCEAVAFDAANGKLLPFQVLSSRKRACASAEDVRVPVCVFVFDLLYLNGAPLVGSTLAHRRKLLREHFRTQPGTWQFATSKDCSTIEEVQLFLEEAVKESCEGLMVKMLTGDNSKYDIARRSHNWLKLKKDYLEGVGDTVDAVIIGAYRGKGKRAGVYGGFLVACYDPESEQFQSLCKLGTGFSDEDLQALSDMLKEHIIDSPRNYYNYDSSHSADVWTSAAVVVEVRCADLSLSPAHRAAIARLHPHKGVSLRFPRFVRVREDKAAEQATTAAQVAQLYLAQDQIKNQVKQPDAAHEDDFY
Summary
Catalytic Activity
ATP + (deoxyribonucleotide)(n)-3'-hydroxyl + 5'-phospho-(deoxyribonucleotide)(m) = (deoxyribonucleotide)(n+m) + AMP + diphosphate.
Cofactor
Mg(2+)
Similarity
Belongs to the ATP-dependent DNA ligase family.
Belongs to the ubiquitin-activating E1 family.
Belongs to the ETS family.
Belongs to the ubiquitin-activating E1 family.
Belongs to the ETS family.
Keywords
ATP-binding
Cell cycle
Cell division
DNA damage
DNA recombination
DNA repair
DNA replication
Ligase
Magnesium
Metal-binding
Nucleotide-binding
Nucleus
Feature
chain DNA ligase 1
Uniprot
A0A2W1BRB8
A0A0N1IDS8
A0A194QBR1
A0A310SSJ1
A0A3L8E1A5
A0A0J7L6A6
+ More
A0A0L7RIX0 A0A026WD56 A0A0C9R7A2 A0A1B0GKD1 A0A088AVT0 E9IA98 E2A154 A0A0P4VZ05 A0A1B6DEE5 A0A0M8ZRH0 A0A1Y1N860 A0A1Y1N567 A0A069DZK5 T1HIR0 E2B772 A0A2J7QBH5 A0A158NEQ8 A0A023F491 A0A067RJ93 A0A2J7QBF6 A0A2J7QBF7 A0A224X8E2 A0A2J7QBF5 A0A195E099 F4WRN8 A0A2J7QBG1 A0A195FMY6 A0A0V0G787 A0A2A3EPL3 A0A195AVS3 A0A232F4R4 A0A1W4WY29 A0A1W4WNC1 K7IZI0 A0A1B0D270 A0A195CRA6 A0A151XIL7 A0A1L8FMV1 A0A210PFJ6 D6WF20 V9I8Y1 A0A2H8TVL3 A0A2R7VVH4 P51892 A0A2R8Q0Q5 B0S533 A0A154PPG8 F6WCY3 U5EV64 J9K4X4 A0A087U1W4 A0A3R7MMP4 A0A182IU67 A0A098LZ92 A0A084W4E8 J9HJ51 A0A182N7T5 H9GHC9 A0A1S4G6J7 W5JIY5 A0A2Y9D1S0 A0A444U0M2 A0A402FQ34 A0A1Q3FW15 A0A3B4CXZ5 A0A2S2NAI6 A0A182RG56 A0A182WHA2 K7FZW6 A0A182PAL8 A0A182XKU0 A0A3N0XH30 A0A1S4H0H7 K7FZX9 A0A452GH34 A0A182I5A2 A0A182H3P9 A0A1S3JWW3 W5M103 A0A3B4EXX9 A0A3P8NLU6 A0A3P9DCC4 Q7PVP5 A0A336MB82 A0A3Q2V7J0 A0A3P8NLY1 A0A2A4J2H1 A0A293LP81 I3KD07 A0A1A7Y584 A0A3B3T509 I3KD06 A0A3Q3GHB0
A0A0L7RIX0 A0A026WD56 A0A0C9R7A2 A0A1B0GKD1 A0A088AVT0 E9IA98 E2A154 A0A0P4VZ05 A0A1B6DEE5 A0A0M8ZRH0 A0A1Y1N860 A0A1Y1N567 A0A069DZK5 T1HIR0 E2B772 A0A2J7QBH5 A0A158NEQ8 A0A023F491 A0A067RJ93 A0A2J7QBF6 A0A2J7QBF7 A0A224X8E2 A0A2J7QBF5 A0A195E099 F4WRN8 A0A2J7QBG1 A0A195FMY6 A0A0V0G787 A0A2A3EPL3 A0A195AVS3 A0A232F4R4 A0A1W4WY29 A0A1W4WNC1 K7IZI0 A0A1B0D270 A0A195CRA6 A0A151XIL7 A0A1L8FMV1 A0A210PFJ6 D6WF20 V9I8Y1 A0A2H8TVL3 A0A2R7VVH4 P51892 A0A2R8Q0Q5 B0S533 A0A154PPG8 F6WCY3 U5EV64 J9K4X4 A0A087U1W4 A0A3R7MMP4 A0A182IU67 A0A098LZ92 A0A084W4E8 J9HJ51 A0A182N7T5 H9GHC9 A0A1S4G6J7 W5JIY5 A0A2Y9D1S0 A0A444U0M2 A0A402FQ34 A0A1Q3FW15 A0A3B4CXZ5 A0A2S2NAI6 A0A182RG56 A0A182WHA2 K7FZW6 A0A182PAL8 A0A182XKU0 A0A3N0XH30 A0A1S4H0H7 K7FZX9 A0A452GH34 A0A182I5A2 A0A182H3P9 A0A1S3JWW3 W5M103 A0A3B4EXX9 A0A3P8NLU6 A0A3P9DCC4 Q7PVP5 A0A336MB82 A0A3Q2V7J0 A0A3P8NLY1 A0A2A4J2H1 A0A293LP81 I3KD07 A0A1A7Y584 A0A3B3T509 I3KD06 A0A3Q3GHB0
EC Number
6.5.1.1
Pubmed
28756777
26354079
30249741
24508170
21282665
20798317
+ More
27129103 28004739 26334808 21347285 25474469 24845553 21719571 28648823 20075255 27762356 28812685 18362917 19820115 8682316 23594743 20431018 24438588 17510324 21881562 20920257 23761445 17381049 12364791 28562605 26483478 25186727 29240929
27129103 28004739 26334808 21347285 25474469 24845553 21719571 28648823 20075255 27762356 28812685 18362917 19820115 8682316 23594743 20431018 24438588 17510324 21881562 20920257 23761445 17381049 12364791 28562605 26483478 25186727 29240929
EMBL
KZ149926
PZC77588.1
KQ461072
KPJ09864.1
KQ459193
KPJ02993.1
+ More
KQ759817 OAD62767.1 QOIP01000001 RLU26447.1 LBMM01000509 KMQ98186.1 KQ414582 KOC70892.1 KK107260 EZA54005.1 GBYB01003825 GBYB01011856 JAG73592.1 JAG81623.1 AJWK01028672 GL761997 EFZ22486.1 GL435707 EFN72821.1 GDKW01000754 JAI55841.1 GEDC01013234 JAS24064.1 KQ435922 KOX68476.1 GEZM01012247 GEZM01012246 GEZM01012243 GEZM01012242 JAV93070.1 GEZM01012249 GEZM01012248 GEZM01012245 GEZM01012244 JAV93051.1 GBGD01000480 JAC88409.1 ACPB03007889 GL446149 EFN88433.1 NEVH01016298 PNF25919.1 ADTU01013610 ADTU01013611 GBBI01002760 JAC15952.1 KK852601 KDR20511.1 PNF25917.1 PNF25918.1 GFTR01007810 JAW08616.1 PNF25915.1 KQ979999 KYN18304.1 GL888292 EGI63045.1 PNF25920.1 KQ981490 KYN41274.1 GECL01002548 JAP03576.1 KZ288198 PBC33657.1 KQ976736 KYM76125.1 NNAY01000947 OXU25784.1 AJVK01010498 KQ977381 KYN03170.1 KQ982080 KYQ60246.1 CM004478 OCT72895.1 NEDP02076736 OWF35231.1 KQ971326 EFA01343.2 JR035926 JR035932 AEY56794.1 AEY56799.1 GFXV01006036 MBW17841.1 KK854113 PTY11497.1 L43496 BX005264 BX294126 BX005060 KQ434998 KZC13334.1 AAMC01120211 GANO01003548 JAB56323.1 ABLF02020542 KK117759 KFM71353.1 QCYY01003062 ROT65649.1 GBSI01000743 JAC95753.1 ATLV01020328 KE525298 KFB45092.1 CH477719 EJY57894.1 AAWZ02022076 ADMH02001277 ETN63253.1 SCEB01215590 RXM28714.1 BDOT01000435 GCF58589.1 GFDL01003403 JAV31642.1 GGMR01001540 MBY14159.1 AGCU01152702 AGCU01152703 RJVU01073245 ROI24873.1 AAAB01008984 APCN01003840 JXUM01107754 JXUM01107755 KQ565171 KXJ71253.1 AHAT01038862 AHAT01038863 AHAT01038864 AHAT01038865 AHAT01038866 EAA15024.3 UFQT01000690 SSX26591.1 NWSH01003682 PCG65936.1 GFWV01003920 MAA28650.1 AERX01017771 AERX01017772 HADW01014232 HADX01003209 SBP25441.1
KQ759817 OAD62767.1 QOIP01000001 RLU26447.1 LBMM01000509 KMQ98186.1 KQ414582 KOC70892.1 KK107260 EZA54005.1 GBYB01003825 GBYB01011856 JAG73592.1 JAG81623.1 AJWK01028672 GL761997 EFZ22486.1 GL435707 EFN72821.1 GDKW01000754 JAI55841.1 GEDC01013234 JAS24064.1 KQ435922 KOX68476.1 GEZM01012247 GEZM01012246 GEZM01012243 GEZM01012242 JAV93070.1 GEZM01012249 GEZM01012248 GEZM01012245 GEZM01012244 JAV93051.1 GBGD01000480 JAC88409.1 ACPB03007889 GL446149 EFN88433.1 NEVH01016298 PNF25919.1 ADTU01013610 ADTU01013611 GBBI01002760 JAC15952.1 KK852601 KDR20511.1 PNF25917.1 PNF25918.1 GFTR01007810 JAW08616.1 PNF25915.1 KQ979999 KYN18304.1 GL888292 EGI63045.1 PNF25920.1 KQ981490 KYN41274.1 GECL01002548 JAP03576.1 KZ288198 PBC33657.1 KQ976736 KYM76125.1 NNAY01000947 OXU25784.1 AJVK01010498 KQ977381 KYN03170.1 KQ982080 KYQ60246.1 CM004478 OCT72895.1 NEDP02076736 OWF35231.1 KQ971326 EFA01343.2 JR035926 JR035932 AEY56794.1 AEY56799.1 GFXV01006036 MBW17841.1 KK854113 PTY11497.1 L43496 BX005264 BX294126 BX005060 KQ434998 KZC13334.1 AAMC01120211 GANO01003548 JAB56323.1 ABLF02020542 KK117759 KFM71353.1 QCYY01003062 ROT65649.1 GBSI01000743 JAC95753.1 ATLV01020328 KE525298 KFB45092.1 CH477719 EJY57894.1 AAWZ02022076 ADMH02001277 ETN63253.1 SCEB01215590 RXM28714.1 BDOT01000435 GCF58589.1 GFDL01003403 JAV31642.1 GGMR01001540 MBY14159.1 AGCU01152702 AGCU01152703 RJVU01073245 ROI24873.1 AAAB01008984 APCN01003840 JXUM01107754 JXUM01107755 KQ565171 KXJ71253.1 AHAT01038862 AHAT01038863 AHAT01038864 AHAT01038865 AHAT01038866 EAA15024.3 UFQT01000690 SSX26591.1 NWSH01003682 PCG65936.1 GFWV01003920 MAA28650.1 AERX01017771 AERX01017772 HADW01014232 HADX01003209 SBP25441.1
Proteomes
UP000053240
UP000053268
UP000279307
UP000036403
UP000053825
UP000053097
+ More
UP000092461 UP000005203 UP000000311 UP000053105 UP000015103 UP000008237 UP000235965 UP000005205 UP000027135 UP000078492 UP000007755 UP000078541 UP000242457 UP000078540 UP000215335 UP000192223 UP000002358 UP000092462 UP000078542 UP000075809 UP000186698 UP000242188 UP000007266 UP000000437 UP000076502 UP000008143 UP000007819 UP000054359 UP000283509 UP000075880 UP000030765 UP000008820 UP000075884 UP000001646 UP000000673 UP000075900 UP000288954 UP000261440 UP000075920 UP000007267 UP000075885 UP000076407 UP000291020 UP000075840 UP000069940 UP000249989 UP000085678 UP000018468 UP000261460 UP000265100 UP000265160 UP000007062 UP000264840 UP000218220 UP000005207 UP000261540 UP000264800
UP000092461 UP000005203 UP000000311 UP000053105 UP000015103 UP000008237 UP000235965 UP000005205 UP000027135 UP000078492 UP000007755 UP000078541 UP000242457 UP000078540 UP000215335 UP000192223 UP000002358 UP000092462 UP000078542 UP000075809 UP000186698 UP000242188 UP000007266 UP000000437 UP000076502 UP000008143 UP000007819 UP000054359 UP000283509 UP000075880 UP000030765 UP000008820 UP000075884 UP000001646 UP000000673 UP000075900 UP000288954 UP000261440 UP000075920 UP000007267 UP000075885 UP000076407 UP000291020 UP000075840 UP000069940 UP000249989 UP000085678 UP000018468 UP000261460 UP000265100 UP000265160 UP000007062 UP000264840 UP000218220 UP000005207 UP000261540 UP000264800
Pfam
Interpro
IPR016059
DNA_ligase_ATP-dep_CS
+ More
IPR012340 NA-bd_OB-fold
IPR012310 DNA_ligase_ATP-dep_cent
IPR012308 DNA_ligase_ATP-dep_N
IPR000977 DNA_ligase_ATP-dep
IPR012309 DNA_ligase_ATP-dep_C
IPR036599 DNA_ligase_N_sf
IPR032420 E1_4HB
IPR042063 Ubi_acti_E1_SCCH
IPR018074 UBQ-activ_enz_E1_CS
IPR042302 E1_FCCH_sf
IPR032418 E1_FCCH
IPR033127 UBQ-activ_enz_E1_Cys_AS
IPR018965 Ub-activating_enz_E1_C
IPR000011 UBQ/SUMO-activ_enz_E1-like
IPR019572 UBA_E1_Cys
IPR000594 ThiF_NAD_FAD-bd
IPR038252 UBA_E1_C_sf
IPR035985 Ubiquitin-activating_enz
IPR018075 UBQ-activ_enz_E1
IPR006073 GTP_binding_domain
IPR027417 P-loop_NTPase
IPR030388 G_ERA_dom
IPR005225 Small_GTP-bd_dom
IPR010497 Epoxide_hydro_N
IPR029058 AB_hydrolase
IPR000639 Epox_hydrolase-like
IPR016482 SecG/Sec61-beta/Sbh
IPR036390 WH_DNA-bd_sf
IPR036388 WH-like_DNA-bd_sf
IPR000418 Ets_dom
IPR012340 NA-bd_OB-fold
IPR012310 DNA_ligase_ATP-dep_cent
IPR012308 DNA_ligase_ATP-dep_N
IPR000977 DNA_ligase_ATP-dep
IPR012309 DNA_ligase_ATP-dep_C
IPR036599 DNA_ligase_N_sf
IPR032420 E1_4HB
IPR042063 Ubi_acti_E1_SCCH
IPR018074 UBQ-activ_enz_E1_CS
IPR042302 E1_FCCH_sf
IPR032418 E1_FCCH
IPR033127 UBQ-activ_enz_E1_Cys_AS
IPR018965 Ub-activating_enz_E1_C
IPR000011 UBQ/SUMO-activ_enz_E1-like
IPR019572 UBA_E1_Cys
IPR000594 ThiF_NAD_FAD-bd
IPR038252 UBA_E1_C_sf
IPR035985 Ubiquitin-activating_enz
IPR018075 UBQ-activ_enz_E1
IPR006073 GTP_binding_domain
IPR027417 P-loop_NTPase
IPR030388 G_ERA_dom
IPR005225 Small_GTP-bd_dom
IPR010497 Epoxide_hydro_N
IPR029058 AB_hydrolase
IPR000639 Epox_hydrolase-like
IPR016482 SecG/Sec61-beta/Sbh
IPR036390 WH_DNA-bd_sf
IPR036388 WH-like_DNA-bd_sf
IPR000418 Ets_dom
SUPFAM
CDD
ProteinModelPortal
A0A2W1BRB8
A0A0N1IDS8
A0A194QBR1
A0A310SSJ1
A0A3L8E1A5
A0A0J7L6A6
+ More
A0A0L7RIX0 A0A026WD56 A0A0C9R7A2 A0A1B0GKD1 A0A088AVT0 E9IA98 E2A154 A0A0P4VZ05 A0A1B6DEE5 A0A0M8ZRH0 A0A1Y1N860 A0A1Y1N567 A0A069DZK5 T1HIR0 E2B772 A0A2J7QBH5 A0A158NEQ8 A0A023F491 A0A067RJ93 A0A2J7QBF6 A0A2J7QBF7 A0A224X8E2 A0A2J7QBF5 A0A195E099 F4WRN8 A0A2J7QBG1 A0A195FMY6 A0A0V0G787 A0A2A3EPL3 A0A195AVS3 A0A232F4R4 A0A1W4WY29 A0A1W4WNC1 K7IZI0 A0A1B0D270 A0A195CRA6 A0A151XIL7 A0A1L8FMV1 A0A210PFJ6 D6WF20 V9I8Y1 A0A2H8TVL3 A0A2R7VVH4 P51892 A0A2R8Q0Q5 B0S533 A0A154PPG8 F6WCY3 U5EV64 J9K4X4 A0A087U1W4 A0A3R7MMP4 A0A182IU67 A0A098LZ92 A0A084W4E8 J9HJ51 A0A182N7T5 H9GHC9 A0A1S4G6J7 W5JIY5 A0A2Y9D1S0 A0A444U0M2 A0A402FQ34 A0A1Q3FW15 A0A3B4CXZ5 A0A2S2NAI6 A0A182RG56 A0A182WHA2 K7FZW6 A0A182PAL8 A0A182XKU0 A0A3N0XH30 A0A1S4H0H7 K7FZX9 A0A452GH34 A0A182I5A2 A0A182H3P9 A0A1S3JWW3 W5M103 A0A3B4EXX9 A0A3P8NLU6 A0A3P9DCC4 Q7PVP5 A0A336MB82 A0A3Q2V7J0 A0A3P8NLY1 A0A2A4J2H1 A0A293LP81 I3KD07 A0A1A7Y584 A0A3B3T509 I3KD06 A0A3Q3GHB0
A0A0L7RIX0 A0A026WD56 A0A0C9R7A2 A0A1B0GKD1 A0A088AVT0 E9IA98 E2A154 A0A0P4VZ05 A0A1B6DEE5 A0A0M8ZRH0 A0A1Y1N860 A0A1Y1N567 A0A069DZK5 T1HIR0 E2B772 A0A2J7QBH5 A0A158NEQ8 A0A023F491 A0A067RJ93 A0A2J7QBF6 A0A2J7QBF7 A0A224X8E2 A0A2J7QBF5 A0A195E099 F4WRN8 A0A2J7QBG1 A0A195FMY6 A0A0V0G787 A0A2A3EPL3 A0A195AVS3 A0A232F4R4 A0A1W4WY29 A0A1W4WNC1 K7IZI0 A0A1B0D270 A0A195CRA6 A0A151XIL7 A0A1L8FMV1 A0A210PFJ6 D6WF20 V9I8Y1 A0A2H8TVL3 A0A2R7VVH4 P51892 A0A2R8Q0Q5 B0S533 A0A154PPG8 F6WCY3 U5EV64 J9K4X4 A0A087U1W4 A0A3R7MMP4 A0A182IU67 A0A098LZ92 A0A084W4E8 J9HJ51 A0A182N7T5 H9GHC9 A0A1S4G6J7 W5JIY5 A0A2Y9D1S0 A0A444U0M2 A0A402FQ34 A0A1Q3FW15 A0A3B4CXZ5 A0A2S2NAI6 A0A182RG56 A0A182WHA2 K7FZW6 A0A182PAL8 A0A182XKU0 A0A3N0XH30 A0A1S4H0H7 K7FZX9 A0A452GH34 A0A182I5A2 A0A182H3P9 A0A1S3JWW3 W5M103 A0A3B4EXX9 A0A3P8NLU6 A0A3P9DCC4 Q7PVP5 A0A336MB82 A0A3Q2V7J0 A0A3P8NLY1 A0A2A4J2H1 A0A293LP81 I3KD07 A0A1A7Y584 A0A3B3T509 I3KD06 A0A3Q3GHB0
PDB
1X9N
E-value=0,
Score=1656
Ontologies
PATHWAY
GO
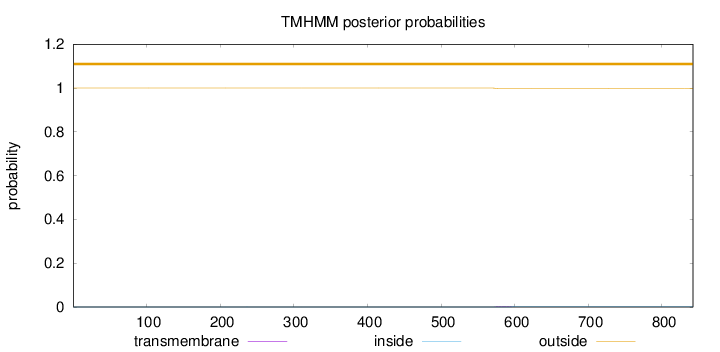
Topology
Subcellular location
Nucleus
Length:
842
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05324
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00037
outside
1 - 842
Population Genetic Test Statistics
Pi
18.139182
Theta
15.785421
Tajima's D
0.05359
CLR
1.202012
CSRT
0.390080495975201
Interpretation
Uncertain
