Pre Gene Modal
BGIBMGA009173
Annotation
PREDICTED:_non-canonical_poly(A)_RNA_polymerase_PAPD5-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.352 Mitochondrial Reliability : 1.094
Sequence
CDS
ATGGATCCCGCCGTCGCGTGGTATCAGCCCGAGCAGGAAGGACCCGCGAAGCGCCTGTGGCTTCGTATTTGGGATACTCAAAGTGAATTAGACAAAATGAACTTGAAAAACATAGAAAACGCAAACCCTAATGTGAACAAAGCACAGGACTTTATACCAATAAATGACGTGAATGGTGAAAATAGGAATAATAATTATTTTAATCCCGCGCGGAGGAAAGTTAACGACAATCGAGCCTCAACTTTTAACCTGAATAAAAATCACGATGCTCTCATAGGAGAATACGGAGGCTGTCCGTGGCGAATACCTAATTATGCGTACAAGCCGGGAGTGTTGGGGCTCCACGAAGAGATCGATCACTTCTACATGTACATGTCGCCGACGGAGACGGAGCACCTGGTCCGCTCGACCGTGGTGAACAGAATCCGCGCCGCCATCCTGTCGCTCTGGCCCCAGGCCCGGGTCGAGGTGTTCGGTTCATTCCGTACTGGACTGTACCTGCCGACCAGCGATATAGACCTAGTCGTTATTGGTGAGTGTCACGGAACGTAG
Protein
MDPAVAWYQPEQEGPAKRLWLRIWDTQSELDKMNLKNIENANPNVNKAQDFIPINDVNGENRNNNYFNPARRKVNDNRASTFNLNKNHDALIGEYGGCPWRIPNYAYKPGVLGLHEEIDHFYMYMSPTETEHLVRSTVVNRIRAAILSLWPQARVEVFGSFRTGLYLPTSDIDLVVIGECHGT
Summary
Uniprot
A0A0L7LFI4
A0A2W1BWL5
A0A2H1VDA3
S4PLT6
A0A194QC43
A0A212FE62
+ More
A0A087ZVN8 A0A2A3E8D7 A0A310SH60 A0A154P4M6 A0A1Y1N563 A0A0L7RFX3 A0A2J7QJQ2 D6WSC3 A0A2J7QJN7 A0A2J7QJP9 V5GIW6 A0A232FDM7 A0A0C9R9K8 A0A0P4VSF2 A0A1Y1NAG4 U5ETV5 A0A1B6EUL3 A0A1L8DWN9 A0A067R9L3 A0A131YZC9 L7MIW8 E0VP78 A0A131XC79 A0A023FMY5 A0A1E1XCS9 A0A147BR44 T1J4Z7 A0A131XXI1 A0A1J1HY03 A0A1B6C358 A0A1B6CKE7 A0A2R5LLF2 U4U7D2 N6TLV4 A0A182L9L7 V3ZFX5 A0A2C9JPD6 A0A0B7BWA9 A0A069DVY1 A0A293LZF1 T1I747 A0A443RKI2 A0A146KMT5 A0A1Z5KZJ7 A0A0P5CFF9 A0A0A9VX23 A0A210R2D7 A0A443SW46 A0A3S1BUY2 A0A2M3YZB5 A0A0A9W4F7 A0A1S3I2A4 A0A1D2NFC8 A0A0N8D1D1 A0A0P5YRG5 A0A0P4Y9H6 A0A0P5BIG1 A0A0P5ZTL9 A0A0P6GMB6 A0A0N8CQX5 A0A0P6IGU1
A0A087ZVN8 A0A2A3E8D7 A0A310SH60 A0A154P4M6 A0A1Y1N563 A0A0L7RFX3 A0A2J7QJQ2 D6WSC3 A0A2J7QJN7 A0A2J7QJP9 V5GIW6 A0A232FDM7 A0A0C9R9K8 A0A0P4VSF2 A0A1Y1NAG4 U5ETV5 A0A1B6EUL3 A0A1L8DWN9 A0A067R9L3 A0A131YZC9 L7MIW8 E0VP78 A0A131XC79 A0A023FMY5 A0A1E1XCS9 A0A147BR44 T1J4Z7 A0A131XXI1 A0A1J1HY03 A0A1B6C358 A0A1B6CKE7 A0A2R5LLF2 U4U7D2 N6TLV4 A0A182L9L7 V3ZFX5 A0A2C9JPD6 A0A0B7BWA9 A0A069DVY1 A0A293LZF1 T1I747 A0A443RKI2 A0A146KMT5 A0A1Z5KZJ7 A0A0P5CFF9 A0A0A9VX23 A0A210R2D7 A0A443SW46 A0A3S1BUY2 A0A2M3YZB5 A0A0A9W4F7 A0A1S3I2A4 A0A1D2NFC8 A0A0N8D1D1 A0A0P5YRG5 A0A0P4Y9H6 A0A0P5BIG1 A0A0P5ZTL9 A0A0P6GMB6 A0A0N8CQX5 A0A0P6IGU1
Pubmed
EMBL
JTDY01001400
KOB73971.1
KZ149926
PZC77587.1
ODYU01001931
SOQ38818.1
+ More
GAIX01003865 JAA88695.1 KQ459193 KPJ02994.1 AGBW02008964 OWR52055.1 KZ288327 PBC27960.1 KQ769895 OAD52772.1 KQ434808 KZC06168.1 GEZM01013419 JAV92578.1 KQ414599 KOC69755.1 NEVH01013550 PNF28796.1 KQ971352 EFA06387.1 PNF28797.1 PNF28799.1 GALX01006924 JAB61542.1 NNAY01000415 OXU28559.1 GBYB01013099 JAG82866.1 GDRN01101320 JAI58403.1 GEZM01013420 JAV92577.1 GANO01004183 JAB55688.1 GECZ01028201 JAS41568.1 GFDF01003260 JAV10824.1 KK852611 KDR20267.1 GEDV01004707 JAP83850.1 GACK01001811 JAA63223.1 DS235357 EEB15184.1 GEFH01004901 JAP63680.1 GBBK01002237 JAC22245.1 GFAC01002136 JAT97052.1 GEGO01002549 JAR92855.1 JH431850 GEFM01004409 JAP71387.1 CVRI01000036 CRK92967.1 GEDC01029350 JAS07948.1 GEDC01023398 JAS13900.1 GGLE01006217 MBY10343.1 KB631817 ERL86526.1 APGK01059362 APGK01059363 KB741293 ENN70220.1 KB203796 ESO83032.1 HACG01050357 CEK97222.1 GBGD01000903 JAC87986.1 GFWV01008492 MAA33221.1 ACPB03005112 NCKU01000373 NCKU01000367 RWS15748.1 RWS15779.1 GDHC01021584 JAP97044.1 GFJQ02006505 JAW00465.1 GDIP01171088 JAJ52314.1 GBHO01043863 JAF99740.1 NEDP02000756 OWF55137.1 NCKV01000078 RWS31730.1 RQTK01000001 RUS92192.1 GGFM01000861 MBW21612.1 GBHO01043869 GBHO01034799 GBHO01003149 JAF99734.1 JAG08805.1 JAG40455.1 LJIJ01000058 ODN03978.1 GDIP01078399 JAM25316.1 GDIP01093148 GDIP01080955 GDIP01054369 LRGB01001411 JAM49346.1 KZS11989.1 GDIP01232942 JAI90459.1 GDIP01184143 JAJ39259.1 GDIP01043016 JAM60699.1 GDIQ01050873 JAN43864.1 GDIP01107647 JAL96067.1 GDIQ01011368 JAN83369.1
GAIX01003865 JAA88695.1 KQ459193 KPJ02994.1 AGBW02008964 OWR52055.1 KZ288327 PBC27960.1 KQ769895 OAD52772.1 KQ434808 KZC06168.1 GEZM01013419 JAV92578.1 KQ414599 KOC69755.1 NEVH01013550 PNF28796.1 KQ971352 EFA06387.1 PNF28797.1 PNF28799.1 GALX01006924 JAB61542.1 NNAY01000415 OXU28559.1 GBYB01013099 JAG82866.1 GDRN01101320 JAI58403.1 GEZM01013420 JAV92577.1 GANO01004183 JAB55688.1 GECZ01028201 JAS41568.1 GFDF01003260 JAV10824.1 KK852611 KDR20267.1 GEDV01004707 JAP83850.1 GACK01001811 JAA63223.1 DS235357 EEB15184.1 GEFH01004901 JAP63680.1 GBBK01002237 JAC22245.1 GFAC01002136 JAT97052.1 GEGO01002549 JAR92855.1 JH431850 GEFM01004409 JAP71387.1 CVRI01000036 CRK92967.1 GEDC01029350 JAS07948.1 GEDC01023398 JAS13900.1 GGLE01006217 MBY10343.1 KB631817 ERL86526.1 APGK01059362 APGK01059363 KB741293 ENN70220.1 KB203796 ESO83032.1 HACG01050357 CEK97222.1 GBGD01000903 JAC87986.1 GFWV01008492 MAA33221.1 ACPB03005112 NCKU01000373 NCKU01000367 RWS15748.1 RWS15779.1 GDHC01021584 JAP97044.1 GFJQ02006505 JAW00465.1 GDIP01171088 JAJ52314.1 GBHO01043863 JAF99740.1 NEDP02000756 OWF55137.1 NCKV01000078 RWS31730.1 RQTK01000001 RUS92192.1 GGFM01000861 MBW21612.1 GBHO01043869 GBHO01034799 GBHO01003149 JAF99734.1 JAG08805.1 JAG40455.1 LJIJ01000058 ODN03978.1 GDIP01078399 JAM25316.1 GDIP01093148 GDIP01080955 GDIP01054369 LRGB01001411 JAM49346.1 KZS11989.1 GDIP01232942 JAI90459.1 GDIP01184143 JAJ39259.1 GDIP01043016 JAM60699.1 GDIQ01050873 JAN43864.1 GDIP01107647 JAL96067.1 GDIQ01011368 JAN83369.1
Proteomes
UP000037510
UP000053268
UP000007151
UP000005203
UP000242457
UP000076502
+ More
UP000053825 UP000235965 UP000007266 UP000215335 UP000027135 UP000009046 UP000183832 UP000030742 UP000019118 UP000075882 UP000030746 UP000076420 UP000015103 UP000285301 UP000242188 UP000288716 UP000271974 UP000085678 UP000094527 UP000076858
UP000053825 UP000235965 UP000007266 UP000215335 UP000027135 UP000009046 UP000183832 UP000030742 UP000019118 UP000075882 UP000030746 UP000076420 UP000015103 UP000285301 UP000242188 UP000288716 UP000271974 UP000085678 UP000094527 UP000076858
Interpro
ProteinModelPortal
A0A0L7LFI4
A0A2W1BWL5
A0A2H1VDA3
S4PLT6
A0A194QC43
A0A212FE62
+ More
A0A087ZVN8 A0A2A3E8D7 A0A310SH60 A0A154P4M6 A0A1Y1N563 A0A0L7RFX3 A0A2J7QJQ2 D6WSC3 A0A2J7QJN7 A0A2J7QJP9 V5GIW6 A0A232FDM7 A0A0C9R9K8 A0A0P4VSF2 A0A1Y1NAG4 U5ETV5 A0A1B6EUL3 A0A1L8DWN9 A0A067R9L3 A0A131YZC9 L7MIW8 E0VP78 A0A131XC79 A0A023FMY5 A0A1E1XCS9 A0A147BR44 T1J4Z7 A0A131XXI1 A0A1J1HY03 A0A1B6C358 A0A1B6CKE7 A0A2R5LLF2 U4U7D2 N6TLV4 A0A182L9L7 V3ZFX5 A0A2C9JPD6 A0A0B7BWA9 A0A069DVY1 A0A293LZF1 T1I747 A0A443RKI2 A0A146KMT5 A0A1Z5KZJ7 A0A0P5CFF9 A0A0A9VX23 A0A210R2D7 A0A443SW46 A0A3S1BUY2 A0A2M3YZB5 A0A0A9W4F7 A0A1S3I2A4 A0A1D2NFC8 A0A0N8D1D1 A0A0P5YRG5 A0A0P4Y9H6 A0A0P5BIG1 A0A0P5ZTL9 A0A0P6GMB6 A0A0N8CQX5 A0A0P6IGU1
A0A087ZVN8 A0A2A3E8D7 A0A310SH60 A0A154P4M6 A0A1Y1N563 A0A0L7RFX3 A0A2J7QJQ2 D6WSC3 A0A2J7QJN7 A0A2J7QJP9 V5GIW6 A0A232FDM7 A0A0C9R9K8 A0A0P4VSF2 A0A1Y1NAG4 U5ETV5 A0A1B6EUL3 A0A1L8DWN9 A0A067R9L3 A0A131YZC9 L7MIW8 E0VP78 A0A131XC79 A0A023FMY5 A0A1E1XCS9 A0A147BR44 T1J4Z7 A0A131XXI1 A0A1J1HY03 A0A1B6C358 A0A1B6CKE7 A0A2R5LLF2 U4U7D2 N6TLV4 A0A182L9L7 V3ZFX5 A0A2C9JPD6 A0A0B7BWA9 A0A069DVY1 A0A293LZF1 T1I747 A0A443RKI2 A0A146KMT5 A0A1Z5KZJ7 A0A0P5CFF9 A0A0A9VX23 A0A210R2D7 A0A443SW46 A0A3S1BUY2 A0A2M3YZB5 A0A0A9W4F7 A0A1S3I2A4 A0A1D2NFC8 A0A0N8D1D1 A0A0P5YRG5 A0A0P4Y9H6 A0A0P5BIG1 A0A0P5ZTL9 A0A0P6GMB6 A0A0N8CQX5 A0A0P6IGU1
PDB
3NYB
E-value=1.61785e-11,
Score=163
Ontologies
GO
PANTHER
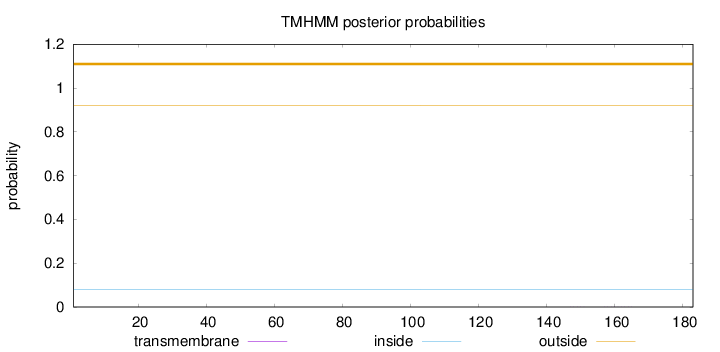
Topology
Length:
183
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00363
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07990
outside
1 - 183
Population Genetic Test Statistics
Pi
2.819918
Theta
18.916379
Tajima's D
-0.870682
CLR
0.942875
CSRT
0.16329183540823
Interpretation
Uncertain
