Gene
KWMTBOMO12321
Pre Gene Modal
BGIBMGA009179
Annotation
PREDICTED:_lipase_1-like_[Bombyx_mori]
Full name
Lipase
Location in the cell
PlasmaMembrane Reliability : 2.429
Sequence
CDS
ATGGCGGCGGGGAGAGCTCACACGCTAAACGTGCTACTCTGGTCCGCGCTCGGAATTATAATCGCGATAAGACTTCCGCTACTGCCGATACGAAATGAGACCAAGCGGCAGTTGGGCTATCCGCCGGACTCCCTGCTGAACGTGACGGAACTAGCGACCCGACACGGGTATCAGTGCGAGGAGCATAACGTAGTCACGGAGGACGGATTCGTGCTGACCGTTTTCCGCGTGTCGAGCGGCCAACGGTGCTCGCGTACACAGATCCCAGTGCTGCTCGTGCACGGTCTGCTGCAGAGCGCTGACTCGTGGCTGGACGCGGGGCCCGGCTCCGGCCTCGCCTACTTGCTGGCAGACGCTTGCCACGACGTGTGGCTCTCCAACACTCGCGGCAACTATTACTCGCGGCGCCACCTCACTCTCGATCCCGACTCAGACGCCGCCTACTGGAACTTCTCAGCCGACGAAATCGGGTACTTTGACTTGCCAGCCATCCTTGATTATGTCCTCAACCGGTCCCGAGCCGACATCCTCAACTACGTGGGCTTCTCGCAGGGCGCGGGAACCTTCTTCATCATGTGTTCCGAGAGACCGAACTATTGTGAAAAGATCAAACTCATGATAGCTTTGGCTCCGGCGACGAGACAGACGAATACGAAATCAACGATGTATCGAAGGCTGACTCAAGCAGTGCAGGGTTGGGAGACGCTGTTGACGACGGCAGGTCTACATGAAATTTTTTCCAAGGGTGCTTTAAGCCAAGAGTTTGTGGCCTTCTTTTGTCACTTCACTAAGTTCACGGAGGGACTTTGCGATGCCGGCCTAAACAAGTTTAACTCTGTCGTTGCTCACCCTGGATCGATGTCAAACGAAACGACAAGAGTGCTCTTTGGACATTTCCCGGCTGGAACATCGCTGCACAACATGGCAAAGTACGGACAGGCAATGACAAATGACAAATTTGTGAAGTTCAACTACGGACATCAGAAGAATTTAGAACTGTACGGAACCTCGGAGCCGCCAGAGTATAACATATCGGCTGTATCACCCCCAGTCGTGGTAATTTACGGGGAGAATGACGGATTAGTGGACACCAAGGATGTTGAATGGGCGATGGAAAGACTCCCAAACGTTCTAGAAGCAGTCCTAATCAAGGACCCGGCTTGGAACCACATGAGTGTTACCTACAGTAGTCGTGTGGAGGAGATAATTTTCCCTAAGATCAATGAGTACCTACAAATATACAGCAGTACGTGA
Protein
MAAGRAHTLNVLLWSALGIIIAIRLPLLPIRNETKRQLGYPPDSLLNVTELATRHGYQCEEHNVVTEDGFVLTVFRVSSGQRCSRTQIPVLLVHGLLQSADSWLDAGPGSGLAYLLADACHDVWLSNTRGNYYSRRHLTLDPDSDAAYWNFSADEIGYFDLPAILDYVLNRSRADILNYVGFSQGAGTFFIMCSERPNYCEKIKLMIALAPATRQTNTKSTMYRRLTQAVQGWETLLTTAGLHEIFSKGALSQEFVAFFCHFTKFTEGLCDAGLNKFNSVVAHPGSMSNETTRVLFGHFPAGTSLHNMAKYGQAMTNDKFVKFNYGHQKNLELYGTSEPPEYNISAVSPPVVVIYGENDGLVDTKDVEWAMERLPNVLEAVLIKDPAWNHMSVTYSSRVEEIIFPKINEYLQIYSST
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Feature
chain Lipase
Uniprot
H9JI31
A0A0N1INH9
A0A1V0M8B4
A0A291P144
A0A194QDF4
A0A088ML86
+ More
A0A0N1PFF0 A0A2W1BV86 A0A2H1W662 A0A2W1BR25 A0A0L7L4B1 A0A2A4J744 A0A2W1BR83 A0A0L7K4K3 A0A0N0PBA5 A0A194QC53 A0A0N1PHZ1 A0A194QBS0 A0A0N0PB17 A0A194QC12 A0A0N1I5B8 A0A0N0PB52 I4DNB5 A0A194QHR9 A0A194QC07 A0A194PSJ3 I4DJ25 A0A194QHS4 A0A0N1IG92 I4DNB6 A0A0N0PB30 A0A0N1IC21 A0A0N1PGW1 A0A212FGL9 I4DRW9 I4DPY2 A0A194QDG0 A0A1L8D695 A0A1L8D6J0 A0A1L8D684 A0A0N1I738 A0A194PRT8 A0A0L7KXY5 A0A194QC17 A0A2W1BMU4 A0A212F4K7 A0A2H1VHW7 A0A2A4K8I4 A0A3F2YX72 A0A182RCW3 A0A182LFG8 Q7QBX7 A0A182XID0 A0A182HTB5 A0A182V7K7 A0A182TPG6 A0A182PA26 A0A182MKD5 A0A182Y799 A0A182GFS9 A0A182SY57 A0A182H3N4 A0A182VQ94 A0A212FB51 B0W003 A0A1W7R7I2 A0A1W7R7P1 B0W6G9 A0A182RXY7 F5GTG5 A0A182FR70 A0A2H1WUD0 A0A182KCP4 A0A2A4K633 A0A1L8D6A4 U5EWU4 A0A182GFT0 A0A1B2M4Y7 B0W6G5 A0A2H1X0E5 W5JF51 A0A2J7QW32 A0A2H1VT20 Q17BM4 B0W6H2 Q16F25 A0A1S4FWC2 A0A1B6C389 Q16MC9 A0A084W6W2 A0A182RXX8 A0A194PL00 A0A1S4G336 B0W6G6 A0A1Q3FSH9 A0A2H1VV71 A0A1S4FVU8 B0W6H1 B4KKR3
A0A0N1PFF0 A0A2W1BV86 A0A2H1W662 A0A2W1BR25 A0A0L7L4B1 A0A2A4J744 A0A2W1BR83 A0A0L7K4K3 A0A0N0PBA5 A0A194QC53 A0A0N1PHZ1 A0A194QBS0 A0A0N0PB17 A0A194QC12 A0A0N1I5B8 A0A0N0PB52 I4DNB5 A0A194QHR9 A0A194QC07 A0A194PSJ3 I4DJ25 A0A194QHS4 A0A0N1IG92 I4DNB6 A0A0N0PB30 A0A0N1IC21 A0A0N1PGW1 A0A212FGL9 I4DRW9 I4DPY2 A0A194QDG0 A0A1L8D695 A0A1L8D6J0 A0A1L8D684 A0A0N1I738 A0A194PRT8 A0A0L7KXY5 A0A194QC17 A0A2W1BMU4 A0A212F4K7 A0A2H1VHW7 A0A2A4K8I4 A0A3F2YX72 A0A182RCW3 A0A182LFG8 Q7QBX7 A0A182XID0 A0A182HTB5 A0A182V7K7 A0A182TPG6 A0A182PA26 A0A182MKD5 A0A182Y799 A0A182GFS9 A0A182SY57 A0A182H3N4 A0A182VQ94 A0A212FB51 B0W003 A0A1W7R7I2 A0A1W7R7P1 B0W6G9 A0A182RXY7 F5GTG5 A0A182FR70 A0A2H1WUD0 A0A182KCP4 A0A2A4K633 A0A1L8D6A4 U5EWU4 A0A182GFT0 A0A1B2M4Y7 B0W6G5 A0A2H1X0E5 W5JF51 A0A2J7QW32 A0A2H1VT20 Q17BM4 B0W6H2 Q16F25 A0A1S4FWC2 A0A1B6C389 Q16MC9 A0A084W6W2 A0A182RXX8 A0A194PL00 A0A1S4G336 B0W6G6 A0A1Q3FSH9 A0A2H1VV71 A0A1S4FVU8 B0W6H1 B4KKR3
Pubmed
EMBL
BABH01040406
BABH01040407
KQ461072
KPJ09860.1
KU755513
ARD71223.1
+ More
MF687644 ATJ44570.1 KQ459193 KPJ03000.1 KJ579224 AIN34700.1 KPJ09861.1 KZ149926 PZC77544.1 ODYU01006566 SOQ48517.1 PZC77549.1 JTDY01003010 KOB70312.1 NWSH01002673 PCG67699.1 PZC77548.1 JTDY01011703 KOB54096.1 KPJ09856.1 KPJ03004.1 KPJ09857.1 KPJ03003.1 KPJ09858.1 KPJ03002.1 KPJ09859.1 KQ461062 KPJ09925.1 AK402824 BAM19405.1 KPJ03001.1 KPJ02997.1 KQ459593 KPI96406.1 AK401293 BAM17915.1 KPJ03006.1 KQ460036 KPJ18427.1 AK402825 BAM19406.1 KPJ09855.1 KPJ09854.1 KPJ09853.1 AGBW02008645 OWR52878.1 AK405305 BAM20659.1 AK403839 BAM19972.1 KPJ03005.1 GEYN01000153 JAV01976.1 GEYN01000165 JAV01964.1 GEYN01000160 JAV01969.1 KPJ09852.1 KQ459596 KPI95464.1 JTDY01004649 KOB67911.1 KPJ03007.1 KZ150017 PZC74945.1 AGBW02010351 OWR48653.1 ODYU01002626 SOQ40371.1 NWSH01000070 PCG79980.1 AXCN02000051 AAAB01008859 EAA08216.4 APCN01000436 AXCM01001786 JXUM01060253 KQ562094 KXJ76706.1 JXUM01107700 KQ565165 KXJ71267.1 AGBW02009374 OWR50972.1 DS231816 EDS37575.1 GEHC01000531 JAV47114.1 GEHC01000530 JAV47115.1 DS231848 EDS36643.1 JI484238 AEB26289.1 ODYU01011128 SOQ56675.1 NWSH01000119 PCG79374.1 GEYN01000147 JAV01982.1 GANO01002812 JAB57059.1 JXUM01060254 KXJ76707.1 KX447604 AOA60271.1 EDS36639.1 ODYU01012460 SOQ58781.1 ADMH02001587 ETN61958.1 NEVH01009768 PNF32784.1 ODYU01004246 SOQ43916.1 CH477320 EAT43642.1 EDS36646.1 CH478517 EAT32845.1 GEDC01029317 GEDC01024313 GEDC01024096 GEDC01020251 JAS07981.1 JAS12985.1 JAS13202.1 JAS17047.1 CH477866 EAT35489.1 ATLV01020990 KE525311 KFB45956.1 KQ459601 KPI94007.1 EDS36640.1 GFDL01004643 JAV30402.1 ODYU01004598 SOQ44646.1 EDS36645.1 CH933807 EDW12727.1 KRG03395.1
MF687644 ATJ44570.1 KQ459193 KPJ03000.1 KJ579224 AIN34700.1 KPJ09861.1 KZ149926 PZC77544.1 ODYU01006566 SOQ48517.1 PZC77549.1 JTDY01003010 KOB70312.1 NWSH01002673 PCG67699.1 PZC77548.1 JTDY01011703 KOB54096.1 KPJ09856.1 KPJ03004.1 KPJ09857.1 KPJ03003.1 KPJ09858.1 KPJ03002.1 KPJ09859.1 KQ461062 KPJ09925.1 AK402824 BAM19405.1 KPJ03001.1 KPJ02997.1 KQ459593 KPI96406.1 AK401293 BAM17915.1 KPJ03006.1 KQ460036 KPJ18427.1 AK402825 BAM19406.1 KPJ09855.1 KPJ09854.1 KPJ09853.1 AGBW02008645 OWR52878.1 AK405305 BAM20659.1 AK403839 BAM19972.1 KPJ03005.1 GEYN01000153 JAV01976.1 GEYN01000165 JAV01964.1 GEYN01000160 JAV01969.1 KPJ09852.1 KQ459596 KPI95464.1 JTDY01004649 KOB67911.1 KPJ03007.1 KZ150017 PZC74945.1 AGBW02010351 OWR48653.1 ODYU01002626 SOQ40371.1 NWSH01000070 PCG79980.1 AXCN02000051 AAAB01008859 EAA08216.4 APCN01000436 AXCM01001786 JXUM01060253 KQ562094 KXJ76706.1 JXUM01107700 KQ565165 KXJ71267.1 AGBW02009374 OWR50972.1 DS231816 EDS37575.1 GEHC01000531 JAV47114.1 GEHC01000530 JAV47115.1 DS231848 EDS36643.1 JI484238 AEB26289.1 ODYU01011128 SOQ56675.1 NWSH01000119 PCG79374.1 GEYN01000147 JAV01982.1 GANO01002812 JAB57059.1 JXUM01060254 KXJ76707.1 KX447604 AOA60271.1 EDS36639.1 ODYU01012460 SOQ58781.1 ADMH02001587 ETN61958.1 NEVH01009768 PNF32784.1 ODYU01004246 SOQ43916.1 CH477320 EAT43642.1 EDS36646.1 CH478517 EAT32845.1 GEDC01029317 GEDC01024313 GEDC01024096 GEDC01020251 JAS07981.1 JAS12985.1 JAS13202.1 JAS17047.1 CH477866 EAT35489.1 ATLV01020990 KE525311 KFB45956.1 KQ459601 KPI94007.1 EDS36640.1 GFDL01004643 JAV30402.1 ODYU01004598 SOQ44646.1 EDS36645.1 CH933807 EDW12727.1 KRG03395.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000218220
UP000007151
+ More
UP000075886 UP000075900 UP000075882 UP000007062 UP000076407 UP000075840 UP000075903 UP000075902 UP000075885 UP000075883 UP000076408 UP000069940 UP000249989 UP000075901 UP000075920 UP000002320 UP000069272 UP000075881 UP000000673 UP000235965 UP000008820 UP000030765 UP000009192
UP000075886 UP000075900 UP000075882 UP000007062 UP000076407 UP000075840 UP000075903 UP000075902 UP000075885 UP000075883 UP000076408 UP000069940 UP000249989 UP000075901 UP000075920 UP000002320 UP000069272 UP000075881 UP000000673 UP000235965 UP000008820 UP000030765 UP000009192
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JI31
A0A0N1INH9
A0A1V0M8B4
A0A291P144
A0A194QDF4
A0A088ML86
+ More
A0A0N1PFF0 A0A2W1BV86 A0A2H1W662 A0A2W1BR25 A0A0L7L4B1 A0A2A4J744 A0A2W1BR83 A0A0L7K4K3 A0A0N0PBA5 A0A194QC53 A0A0N1PHZ1 A0A194QBS0 A0A0N0PB17 A0A194QC12 A0A0N1I5B8 A0A0N0PB52 I4DNB5 A0A194QHR9 A0A194QC07 A0A194PSJ3 I4DJ25 A0A194QHS4 A0A0N1IG92 I4DNB6 A0A0N0PB30 A0A0N1IC21 A0A0N1PGW1 A0A212FGL9 I4DRW9 I4DPY2 A0A194QDG0 A0A1L8D695 A0A1L8D6J0 A0A1L8D684 A0A0N1I738 A0A194PRT8 A0A0L7KXY5 A0A194QC17 A0A2W1BMU4 A0A212F4K7 A0A2H1VHW7 A0A2A4K8I4 A0A3F2YX72 A0A182RCW3 A0A182LFG8 Q7QBX7 A0A182XID0 A0A182HTB5 A0A182V7K7 A0A182TPG6 A0A182PA26 A0A182MKD5 A0A182Y799 A0A182GFS9 A0A182SY57 A0A182H3N4 A0A182VQ94 A0A212FB51 B0W003 A0A1W7R7I2 A0A1W7R7P1 B0W6G9 A0A182RXY7 F5GTG5 A0A182FR70 A0A2H1WUD0 A0A182KCP4 A0A2A4K633 A0A1L8D6A4 U5EWU4 A0A182GFT0 A0A1B2M4Y7 B0W6G5 A0A2H1X0E5 W5JF51 A0A2J7QW32 A0A2H1VT20 Q17BM4 B0W6H2 Q16F25 A0A1S4FWC2 A0A1B6C389 Q16MC9 A0A084W6W2 A0A182RXX8 A0A194PL00 A0A1S4G336 B0W6G6 A0A1Q3FSH9 A0A2H1VV71 A0A1S4FVU8 B0W6H1 B4KKR3
A0A0N1PFF0 A0A2W1BV86 A0A2H1W662 A0A2W1BR25 A0A0L7L4B1 A0A2A4J744 A0A2W1BR83 A0A0L7K4K3 A0A0N0PBA5 A0A194QC53 A0A0N1PHZ1 A0A194QBS0 A0A0N0PB17 A0A194QC12 A0A0N1I5B8 A0A0N0PB52 I4DNB5 A0A194QHR9 A0A194QC07 A0A194PSJ3 I4DJ25 A0A194QHS4 A0A0N1IG92 I4DNB6 A0A0N0PB30 A0A0N1IC21 A0A0N1PGW1 A0A212FGL9 I4DRW9 I4DPY2 A0A194QDG0 A0A1L8D695 A0A1L8D6J0 A0A1L8D684 A0A0N1I738 A0A194PRT8 A0A0L7KXY5 A0A194QC17 A0A2W1BMU4 A0A212F4K7 A0A2H1VHW7 A0A2A4K8I4 A0A3F2YX72 A0A182RCW3 A0A182LFG8 Q7QBX7 A0A182XID0 A0A182HTB5 A0A182V7K7 A0A182TPG6 A0A182PA26 A0A182MKD5 A0A182Y799 A0A182GFS9 A0A182SY57 A0A182H3N4 A0A182VQ94 A0A212FB51 B0W003 A0A1W7R7I2 A0A1W7R7P1 B0W6G9 A0A182RXY7 F5GTG5 A0A182FR70 A0A2H1WUD0 A0A182KCP4 A0A2A4K633 A0A1L8D6A4 U5EWU4 A0A182GFT0 A0A1B2M4Y7 B0W6G5 A0A2H1X0E5 W5JF51 A0A2J7QW32 A0A2H1VT20 Q17BM4 B0W6H2 Q16F25 A0A1S4FWC2 A0A1B6C389 Q16MC9 A0A084W6W2 A0A182RXX8 A0A194PL00 A0A1S4G336 B0W6G6 A0A1Q3FSH9 A0A2H1VV71 A0A1S4FVU8 B0W6H1 B4KKR3
PDB
1K8Q
E-value=2.28568e-52,
Score=520
Ontologies
GO
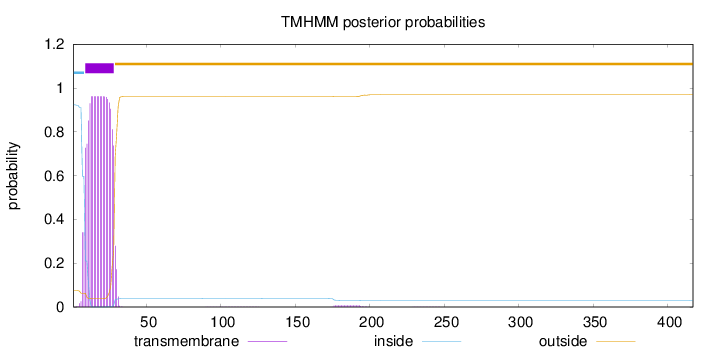
Topology
Length:
417
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.57099
Exp number, first 60 AAs:
19.39985
Total prob of N-in:
0.92449
POSSIBLE N-term signal
sequence
inside
1 - 8
TMhelix
9 - 28
outside
29 - 417
Population Genetic Test Statistics
Pi
29.473886
Theta
23.241649
Tajima's D
0.779802
CLR
1.002871
CSRT
0.5981200939953
Interpretation
Uncertain
