Gene
KWMTBOMO12318
Pre Gene Modal
BGIBMGA009176
Annotation
PREDICTED:_uncharacterized_protein_LOC101738532_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.85
Sequence
CDS
ATGGGCAGCAGCACGACCTCGGACAAGACCATCAACACGAGCACTGATTCAGCGGCCACTGCCCTCACTTCCATCACGGGAACAGATGATTCCATGGACGCTGAGCCTGAACCTCCCGAGCTACCACACAACCCTCCAAGCCTCATCGACGCGGAACCTATCCAAAACCCTCCAAGCACCGCCCCGCCACATTACCACAGACCCCAAAATAGCTGTGACATCGACTCCCCTGCTTACACAAAACCTCAGTCGGCCACCGAGGTCCTAGACTCCCCAAGGCCAGGCCCAGCCCGTTTCATTTTTGACGTTCCACTTCCAGATCCTCGAAAAGCAGATAAAACGCGTAACAAAACCCAAGTGACACAGAGGAGAGGCAGTGAACCTCGCGTGAAAAAGAAAGAGAAAGAGTATTACAAGGACGAAGTGGCCAAGTTACGGAGACAGTTGGAAGGGACGGAAGTCTTTGCGACTAGCAGAAGAAGATCTTCGACGCACCAACATTCTGTTCCACCGCCGTCTTCTAGGAGACAGTCCACAGCAGCCCAGGTCCACGTCACAGCCAACCCGTTGGACTCAGCAGCTACCACCATCCCAGCCGGCAGGACCACCATCGTAGTCCAACAACCATCTCTGTCATTGGACTACGGAGGGTGCTCCACGTTCCTAGTTAGAGAAGGCGAAGATGGAAGGAGAAAAAGTAGGATCTGCACTGATCACCTGCAACAACTCCTTGACGTGACCAGCCAGTTCACGTCGGAAGACTTGCATGAGTTCGACAAGAGATACGGAAGTCCACATCACTCGAGATCCCAGTCAGTGAAGGCCCAAAGATCGAGAACGGGAGAACGACAGCTCCTTGGTCTGCCACAGCAGAGAGCCAGGGTGGCTTCCATGCCTAACACGGGAGTGGAAGAAGAATACTACAGGCTTCGACATTTCAGTATAACTGGTAAGGGCGTAGTGAACCGCGGTGACTCCTTGCGCTCCCGAAGGTCTCGTTCCAACACGTCGGTGGCCTCCAGCGCTTCTAGTACCGAGGTCGCAAAACGAGCCTCAGCGCCCTGCTCCTTGGCATCATCCAGGGACTCATCAGCCGGTCTCAGTTCCTACAGAGTGCTGGTGTTAGGAGGTCCTGGAGTCGGAAAGTCGAGTCTGGTGGGACAGTTCATGACTTCGGAATATCTTCATGCGTACGACACTAGCATAGATGACGAATCTGGCGAGAGATCTGTCTGCGTGCTACTTGCTGGAGAGGAATCCGAGATAACCTTTCTGGATTCGCCACCAGATCATGTAGTGTCCGAAGGTTGTGCCCACGGGTACTGCGTGGTTTACTCCACGGCAGACAGGAGCAGTTTCGTGGAAGCTGAGAGGAGATTGCAGGTTCTGTGGGCCGCTGGACACACCTCGCGCCGTGCTGTCATCCTGGTCGGAAATAAAGCAGATCTGGCGAGATCAAGGGTCGTACCTACTGATGAGGGCAAGAGCTTGGCTACATCTTACGACTGCAAGTTCATTGAGACGTCTGTCGGTATCAATCACAACGTGGACGAGCTGCTGGTCGGACTCCTCACTCAGATCCGACTCAAGCAACAGCACGCCGAGAAGGCCAAAAAACGCAGCGGCTCACGCAAGAACAGAAGCCGCGCCAGGTCTCCGATGGAGAGCGAGCAGCCCGCGCCAGCCCCGGCCTCCTCCTCCGCGGCCAGCACGCCCAGGAAGAGGACCAGACTCTCGGCCAGTGTCAAGGTCCGAGGTCTTCTGGGGCGCGTGTGGGCGCGTGACTCCAAGTCGAAGTCGTGCGAGAACCTGCATGTCCTCTAG
Protein
MGSSTTSDKTINTSTDSAATALTSITGTDDSMDAEPEPPELPHNPPSLIDAEPIQNPPSTAPPHYHRPQNSCDIDSPAYTKPQSATEVLDSPRPGPARFIFDVPLPDPRKADKTRNKTQVTQRRGSEPRVKKKEKEYYKDEVAKLRRQLEGTEVFATSRRRSSTHQHSVPPPSSRRQSTAAQVHVTANPLDSAATTIPAGRTTIVVQQPSLSLDYGGCSTFLVREGEDGRRKSRICTDHLQQLLDVTSQFTSEDLHEFDKRYGSPHHSRSQSVKAQRSRTGERQLLGLPQQRARVASMPNTGVEEEYYRLRHFSITGKGVVNRGDSLRSRRSRSNTSVASSASSTEVAKRASAPCSLASSRDSSAGLSSYRVLVLGGPGVGKSSLVGQFMTSEYLHAYDTSIDDESGERSVCVLLAGEESEITFLDSPPDHVVSEGCAHGYCVVYSTADRSSFVEAERRLQVLWAAGHTSRRAVILVGNKADLARSRVVPTDEGKSLATSYDCKFIETSVGINHNVDELLVGLLTQIRLKQQHAEKAKKRSGSRKNRSRARSPMESEQPAPAPASSSAASTPRKRTRLSASVKVRGLLGRVWARDSKSKSCENLHVL
Summary
Uniprot
Pubmed
EMBL
KQ459193
KPJ03007.1
KZ149926
PZC77578.1
KQ461072
KPJ09851.1
+ More
RSAL01000502 RVE41520.1 NWSH01003486 PCG66200.1 JTDY01003010 KOB70310.1 KK107138 EZA57923.1 KQ971354 EFA06026.2 GL448287 EFN84879.1 ADTU01007418 ADTU01007419 ADTU01007420 ADTU01007421 ADTU01007422 KQ982576 KYQ54650.1 QOIP01000002 RLU25497.1 GBXI01001708 JAD12584.1 CM002911 KMY95070.1 AE013599 AAM70848.2 GGFL01000610 MBW64788.1
RSAL01000502 RVE41520.1 NWSH01003486 PCG66200.1 JTDY01003010 KOB70310.1 KK107138 EZA57923.1 KQ971354 EFA06026.2 GL448287 EFN84879.1 ADTU01007418 ADTU01007419 ADTU01007420 ADTU01007421 ADTU01007422 KQ982576 KYQ54650.1 QOIP01000002 RLU25497.1 GBXI01001708 JAD12584.1 CM002911 KMY95070.1 AE013599 AAM70848.2 GGFL01000610 MBW64788.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
PDB
3Q7Q
E-value=7.08214e-24,
Score=276
Ontologies
KEGG
GO
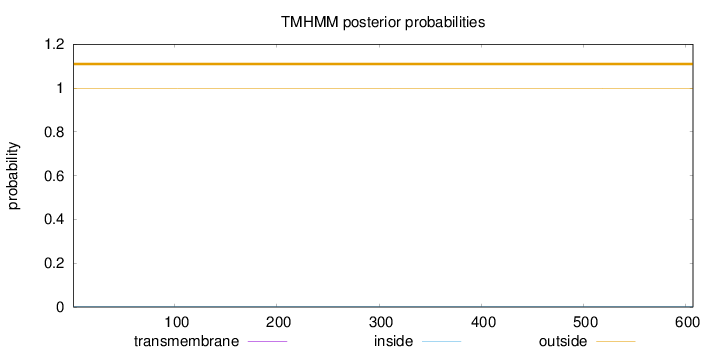
Topology
Length:
607
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00123
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00146
outside
1 - 607
Population Genetic Test Statistics
Pi
25.161658
Theta
24.444584
Tajima's D
-1.601586
CLR
0.896551
CSRT
0.0460476976151192
Interpretation
Uncertain
