Gene
KWMTBOMO12317
Pre Gene Modal
BGIBMGA009178
Annotation
PREDICTED:_uncharacterized_protein_LOC106102902_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 2.303
Sequence
CDS
ATGTATCATCCGGCCAGAAAAAAGTTGATTGATTTTTATTTATCAATTGTTTTAATTATGAGCGTAACAAGGACCGAAGCTAGATCCTTTCAGATGACTGCAAATGCCAAAACGAACGACGATAATTTGGATAATAACTTAATTGATTTCGAAGAAGACAACACATTCAAATTCAACAAAAAGTACGGTATAACGTCGTGGCACGATGACGAATATTTCAATAAAAAAACACACAGATACACTATTGTTAATGACATTGGCGAAAAAATATTCGAAAGTAAAAATAAAGCAAAGAATATTAGAAATAACGATGCTACTACTACAGAAGATTACAGAGTGATCCCACTGGAATTAATTACTACAACTGAGAATACGTTGATAAGTATTATTGAAAGTACTACAGAACAAGATTTGAGTGTTATTCCTTTGTCCACAACCGCAAAAGTACCTAATATAACAATAACAGACTCAATAACATTACCCGTATCTACAGAGTATCCTGAAACTACAACTATTGAAAAAATAGAAATTATTGTACAAGAAAAAGATACCGCCCCAGAAATTTCGAAGGAAGTAGAAGTAGATCTCAACAATGAAACTACTAAATTTCCGTTTGAAACAACAAGTGAGGTTACATTGGAAAGTGATGAAGTTAATGTTTCAACTGAGAGCTTTGATGAAACGACGCTCATAGAACAAATATCGAAAGAAAATTACAACAATTCTAAAGCACTAACTGAAATTTATAAATCTACAGAAGATTCAGTGATTAATACTACAGAAGATACTAATTTTAATGTAACAGATATTCATTTTGAGCTTTTAAACACTACAAAATTGGAAATAAAAAAAACGAATGAAAGCAGAATAGATGAAAGTAGAGAAACCGATGCAGATGTACCAATATTTACAGAATTAGACGCCGAAGAAGAAATAGATGTTCCAGAGGATTATTATGATACTAAAGATGTTGTTCCTACTACGGCTCCAAGGACCGATGCGTTATCAGTTATATTTGGACTTGCAGGGAGCGTTGTGGAGAGTGTAGTAGAGACTGTAGCAGAAAGGGTTGTCCCGAAGAGCGTTTTCGACTTGTTTAAGAGAATGCAGAGGCAGAGCGAAGCTTTAGAATCTGAGCGGCTGAGGAGTAAAGAGGAGAATGGTGGCATAGGTCAATTCACCCGGGGCATCATCAAAACGATATCAACAGGTATCTCGAGACCACTCAGTCAGCTGATGGCTGGAGCTAGAGATATAGGATCTCTGGACACAGATCGGGGATTCGTTTCCAGTTTAGCATCAGGAGTGACCAGTGTTGCCAACGTTGCCAACTCACTTGTCGATACTTTTAAGGATCGGGTGCAAGCTATCTATCCAGGGACGGTGTGGTGTGGCGACGGAAGGTCCGCGGCCGCGCGCTCTGGTGACCTTGGCCTCTTCTTCTTCACGGACACGTGTTGTCGGCAGCACGACGCCTGCAAGCTGTATATCAAGGCTGGAGACTCTAAGTACGGCCTGACCAACACCGGACTCTTCACAAGGTCGCACTGCAGCTGCGACGTGAAGTTCCGGGAGTGTCTGAGGCGCACCAATTCGCTGGTGTCGGCCCAGATAGGGCTGACGTACTTCAACGTGCTGGGGCCGCAGTGCTTCAGGCGGGCTCACCCTATCGTGAGGTGCGCGAGGAGGACGCGCATTACTGGCCAGAGATGCGAAGAGTACGAGTTGGACTTCACCAAGCCCAGGATGTGGCAGTGGTTTGACAACGAGACATTTTAA
Protein
MYHPARKKLIDFYLSIVLIMSVTRTEARSFQMTANAKTNDDNLDNNLIDFEEDNTFKFNKKYGITSWHDDEYFNKKTHRYTIVNDIGEKIFESKNKAKNIRNNDATTTEDYRVIPLELITTTENTLISIIESTTEQDLSVIPLSTTAKVPNITITDSITLPVSTEYPETTTIEKIEIIVQEKDTAPEISKEVEVDLNNETTKFPFETTSEVTLESDEVNVSTESFDETTLIEQISKENYNNSKALTEIYKSTEDSVINTTEDTNFNVTDIHFELLNTTKLEIKKTNESRIDESRETDADVPIFTELDAEEEIDVPEDYYDTKDVVPTTAPRTDALSVIFGLAGSVVESVVETVAERVVPKSVFDLFKRMQRQSEALESERLRSKEENGGIGQFTRGIIKTISTGISRPLSQLMAGARDIGSLDTDRGFVSSLASGVTSVANVANSLVDTFKDRVQAIYPGTVWCGDGRSAAARSGDLGLFFFTDTCCRQHDACKLYIKAGDSKYGLTNTGLFTRSHCSCDVKFRECLRRTNSLVSAQIGLTYFNVLGPQCFRRAHPIVRCARRTRITGQRCEEYELDFTKPRMWQWFDNETF
Summary
Similarity
Belongs to the phospholipase A2 family.
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF05826 Phospholip_A2_2
SUPFAM
SSF48619
SSF48619
Gene 3D
ProteinModelPortal
PDB
1POC
E-value=2.4563e-30,
Score=331
Ontologies
GO
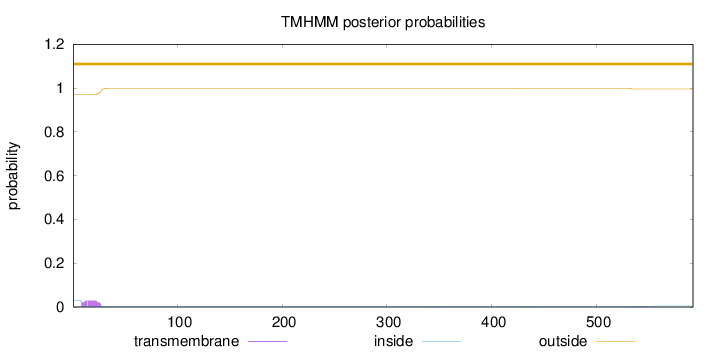
Topology
Subcellular location
Secreted
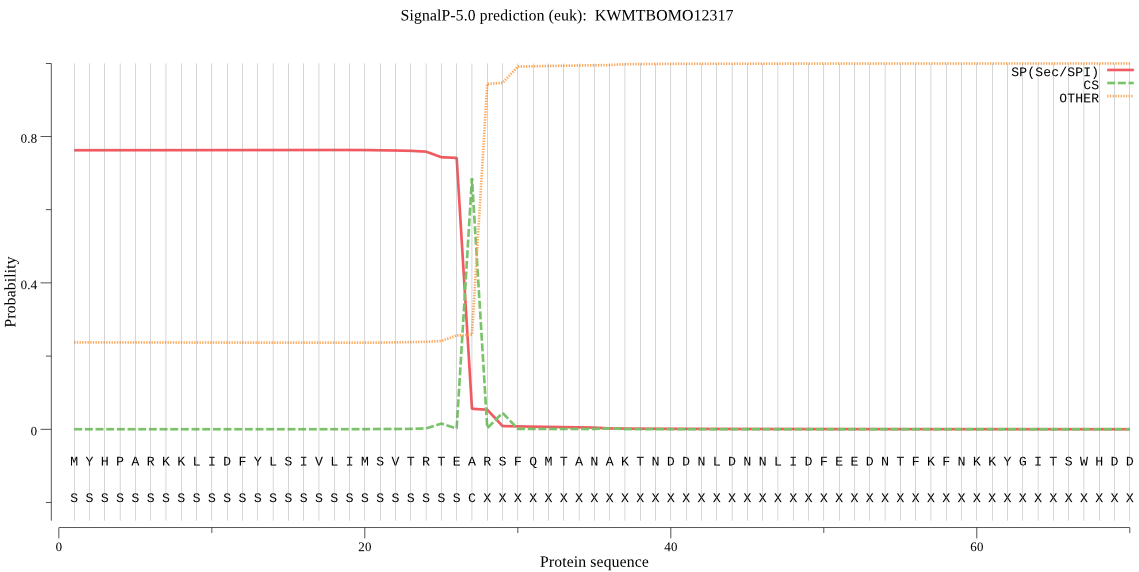
SignalP
Position: 1 - 27,
Likelihood: 0.763487
Length:
592
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.56433
Exp number, first 60 AAs:
0.50536
Total prob of N-in:
0.03031
outside
1 - 592
Population Genetic Test Statistics
Pi
32.228722
Theta
27.952759
Tajima's D
0.483728
CLR
0.685297
CSRT
0.513874306284686
Interpretation
Uncertain
