Gene
KWMTBOMO12312
Pre Gene Modal
BGIBMGA009167
Annotation
PREDICTED:_beta-1?3-galactosyltransferase_1-like_[Bombyx_mori]
Full name
Hexosyltransferase
Location in the cell
PlasmaMembrane Reliability : 3.986
Sequence
CDS
ATGAGGATGACGAATGTATTTTTTTTGACCAATACATTTAGGAAAATAATAATGCGTCGGAGGATTTTTGTAATTCTACCGTTCTTGTTGGGATTGGGAGTCACCATTGTGGCCTGTAGATGGACGGCCGCCGTTGTCAGCCACGTCCTCCTACCACCAGGTCCGCCGGATCAAAACTTGGAGCATTTCATTAGGAATAGAAGTTTAAGTCACTATTTTGATAATATTCAAATCCTCATAGAGCCGTCCGCGCCTTGCCCAGGGTCGGACATTCCTGTGGTAGCGTTGGTAGCTACGACGCCATCACGGTTTGACCAGAGACAAGCCATCAGAGAGACGTGGGCTAAACGTGTGCTTACTTACTTCTTTATGGGCTTGGAAGGAGGGAAAATCGATGAAAGTATGGTAGATATCTACGTAGAAGCCAAGGAACACAGCGATATGGTAATATTGAACTTCCACGAACATTATCAGAATTTAACCTTGAAGACTGGCTTCATGCTGAAGTGGACTCTAGAGCGATGTGCTAACGCCAATTTCCTACTTAAAATCGACGACGATGTCTTCTTAAATCCTTGGAACTTGGAGCAGGTAATGAGGGGAAAGGAACGTCACAAATTGATTGGATACAAAATCAAGGATTCAACTGTACACCGAGATCAGTATACGAAATGGTACTTACCCCGGTGGCTGTATAGCCGTGATGTGATCCCGGAATATCTGGCCGGACCGGCCTATGTCGTAAATACGAAGCACATCGCGGAGATTTTACAGGAAGCAAAGAAGACACCGCTGATCAATTTAGAAGATGTATATTTTACTTATCTAGTCTCCAGGGAAGGTTTGGACTTTGAATTGACTCACGAAAGAACTTTCAGCCCGCACAAGCCCTTCCTGCTGTGCTCATATTGGAGCCTCACGACTGTTCACAGTGTATCTGATACAGAAATGCTAAGCGCTTGGAAAAGTTATAATGACGACGCCAAACAAAATAGTACTTTATGTGGCTTTCGCTTTCCGCCGCTGCGATGA
Protein
MRMTNVFFLTNTFRKIIMRRRIFVILPFLLGLGVTIVACRWTAAVVSHVLLPPGPPDQNLEHFIRNRSLSHYFDNIQILIEPSAPCPGSDIPVVALVATTPSRFDQRQAIRETWAKRVLTYFFMGLEGGKIDESMVDIYVEAKEHSDMVILNFHEHYQNLTLKTGFMLKWTLERCANANFLLKIDDDVFLNPWNLEQVMRGKERHKLIGYKIKDSTVHRDQYTKWYLPRWLYSRDVIPEYLAGPAYVVNTKHIAEILQEAKKTPLINLEDVYFTYLVSREGLDFELTHERTFSPHKPFLLCSYWSLTTVHSVSDTEMLSAWKSYNDDAKQNSTLCGFRFPPLR
Summary
Similarity
Belongs to the glycosyltransferase 31 family.
Feature
chain Hexosyltransferase
Uniprot
H9JI19
A0A2W1C0H6
A0A212ENI5
A0A2H1V461
A0A194QC64
A0A0N1INH7
+ More
A0A2W1BR41 A0A1L8E1E9 A0A1L8E1M0 A0A067QWG9 A0A2P8Y659 A0A1L8E1W0 A0A336M552 A0A1B0CF99 A0A336MN17 A0A2P6LAA8 G3MJ91 A0A1Y1KV48 A0A1Y1KUY2 A0A087YR42 A0A3B3UX26 A0A3B3YZS7 A0A1B0GIK7 A0A2L2Y294 A0A087T2X2 A0A1Y1KZV3 K7EW89 A0A3Q1EGK1 A0A3B5B1H2 A0A1V9X4N4 W4Y0Z9 B7PGF2 A0A3P8TJH5 A0A0K2U674 A0A1B6I5F0 A0A0S7I7T0 A0A3Q1BMB2 A0A1L8F8A2 K7GBA0 A0A2M4ALW2 W5JAY7 A0A087ZCU3 A0A2M4AL69 A0A2R5LKE6 A0A1B6M1H3 A0A3Q7XHS4 A0A2I4AN74 A0A147BCK9 A0A3B4CNK0 A0A224YQ19 A0A2M4BNB4 A0A3Q2PWI1 A0A147AES7 A0A182W5R3 A0A2M4BMR3 A0A2M4BMU1 A0A3B4XW44 A0A3Q2YY68 A0A3B4V7R0 A0A2M3ZKV5 G3VGE5 A0A0F8BC69 A0A3Q1HU30 A0A2U9C2L5 A0A2P6KER0 A0A3P8V1B0 A0A3Q2ZPY1 D2I3E2 A0A3Q2DPH1 A0A3B5A4J4 A0A3Q4AN34 A0A3B4YQC7 G3VBU5 A0A3Q3L7E7 A0A0C9QFA1 A0A0M9ACS9 H2TR38 A0A3Q1F029 A0A3Q1BAD7 A0A3B4U1V3 F1MGN7 A0A1A8DVG6 W5LU59 A0A3B4ZXR7 A0A0C9RIX5 A0A1A8AWB8 L7M312 A0A1A8MEQ5 Q4SDG5 K7E0Y3 A0A1A7XMH4 H2TP59 A0A1S3MWW4
A0A2W1BR41 A0A1L8E1E9 A0A1L8E1M0 A0A067QWG9 A0A2P8Y659 A0A1L8E1W0 A0A336M552 A0A1B0CF99 A0A336MN17 A0A2P6LAA8 G3MJ91 A0A1Y1KV48 A0A1Y1KUY2 A0A087YR42 A0A3B3UX26 A0A3B3YZS7 A0A1B0GIK7 A0A2L2Y294 A0A087T2X2 A0A1Y1KZV3 K7EW89 A0A3Q1EGK1 A0A3B5B1H2 A0A1V9X4N4 W4Y0Z9 B7PGF2 A0A3P8TJH5 A0A0K2U674 A0A1B6I5F0 A0A0S7I7T0 A0A3Q1BMB2 A0A1L8F8A2 K7GBA0 A0A2M4ALW2 W5JAY7 A0A087ZCU3 A0A2M4AL69 A0A2R5LKE6 A0A1B6M1H3 A0A3Q7XHS4 A0A2I4AN74 A0A147BCK9 A0A3B4CNK0 A0A224YQ19 A0A2M4BNB4 A0A3Q2PWI1 A0A147AES7 A0A182W5R3 A0A2M4BMR3 A0A2M4BMU1 A0A3B4XW44 A0A3Q2YY68 A0A3B4V7R0 A0A2M3ZKV5 G3VGE5 A0A0F8BC69 A0A3Q1HU30 A0A2U9C2L5 A0A2P6KER0 A0A3P8V1B0 A0A3Q2ZPY1 D2I3E2 A0A3Q2DPH1 A0A3B5A4J4 A0A3Q4AN34 A0A3B4YQC7 G3VBU5 A0A3Q3L7E7 A0A0C9QFA1 A0A0M9ACS9 H2TR38 A0A3Q1F029 A0A3Q1BAD7 A0A3B4U1V3 F1MGN7 A0A1A8DVG6 W5LU59 A0A3B4ZXR7 A0A0C9RIX5 A0A1A8AWB8 L7M312 A0A1A8MEQ5 Q4SDG5 K7E0Y3 A0A1A7XMH4 H2TP59 A0A1S3MWW4
EC Number
2.4.1.-
Pubmed
EMBL
BABH01035415
BABH01035416
BABH01035417
BABH01035418
KZ149926
PZC77573.1
+ More
AGBW02013665 OWR43011.1 ODYU01000582 SOQ35617.1 KQ459193 KPJ03014.1 KQ461072 KPJ09844.1 PZC77572.1 GFDF01001521 JAV12563.1 GFDF01001520 JAV12564.1 KK852868 KDR14684.1 PYGN01000882 PSN39731.1 GFDF01001522 JAV12562.1 UFQT01000546 SSX25140.1 AJWK01009855 UFQT01001689 SSX31430.1 MWRG01000716 PRD35503.1 JO841942 AEO33559.1 GEZM01073266 JAV65214.1 GEZM01073267 JAV65213.1 AYCK01007049 AJWK01014243 AJWK01014244 AJWK01014245 AJWK01014246 IAAA01006731 LAA02283.1 KK113146 KFM59461.1 GEZM01073265 JAV65215.1 AGCU01071748 MNPL01025622 OQR68222.1 AAGJ04042979 AAGJ04042980 ABJB010038347 DS707924 EEC05674.1 HACA01016372 CDW33733.1 GECU01025553 GECU01017482 GECU01010824 GECU01003970 GECU01002151 JAS82153.1 JAS90224.1 JAS96882.1 JAT03737.1 JAT05556.1 GBYX01432142 GBYX01432141 JAO49172.1 CM004480 OCT67823.1 AGCU01010850 GGFK01008277 MBW41598.1 ADMH02001755 ETN61166.1 GGFK01008212 MBW41533.1 GGLE01005877 MBY10003.1 GEBQ01010233 JAT29744.1 GEGO01006900 JAR88504.1 GFPF01007899 MAA19045.1 GGFJ01005361 MBW54502.1 GCES01009426 JAR76897.1 GGFJ01005235 MBW54376.1 GGFJ01005234 MBW54375.1 GGFM01008381 MBW29132.1 AEFK01088243 KQ041052 KKF31012.1 CP026254 AWP10851.1 MWRG01014020 PRD24815.1 GL194300 EFB23269.1 AEFK01102680 GBYB01013193 JAG82960.1 KQ435700 KOX80499.1 HAEA01008761 SBQ37241.1 GBYB01013192 JAG82959.1 HADY01020286 SBP58771.1 GACK01007555 JAA57479.1 HAEF01013857 SBR55016.1 CAAE01014638 CAG01317.1 HADW01017882 HADX01008529 SBP19282.1
AGBW02013665 OWR43011.1 ODYU01000582 SOQ35617.1 KQ459193 KPJ03014.1 KQ461072 KPJ09844.1 PZC77572.1 GFDF01001521 JAV12563.1 GFDF01001520 JAV12564.1 KK852868 KDR14684.1 PYGN01000882 PSN39731.1 GFDF01001522 JAV12562.1 UFQT01000546 SSX25140.1 AJWK01009855 UFQT01001689 SSX31430.1 MWRG01000716 PRD35503.1 JO841942 AEO33559.1 GEZM01073266 JAV65214.1 GEZM01073267 JAV65213.1 AYCK01007049 AJWK01014243 AJWK01014244 AJWK01014245 AJWK01014246 IAAA01006731 LAA02283.1 KK113146 KFM59461.1 GEZM01073265 JAV65215.1 AGCU01071748 MNPL01025622 OQR68222.1 AAGJ04042979 AAGJ04042980 ABJB010038347 DS707924 EEC05674.1 HACA01016372 CDW33733.1 GECU01025553 GECU01017482 GECU01010824 GECU01003970 GECU01002151 JAS82153.1 JAS90224.1 JAS96882.1 JAT03737.1 JAT05556.1 GBYX01432142 GBYX01432141 JAO49172.1 CM004480 OCT67823.1 AGCU01010850 GGFK01008277 MBW41598.1 ADMH02001755 ETN61166.1 GGFK01008212 MBW41533.1 GGLE01005877 MBY10003.1 GEBQ01010233 JAT29744.1 GEGO01006900 JAR88504.1 GFPF01007899 MAA19045.1 GGFJ01005361 MBW54502.1 GCES01009426 JAR76897.1 GGFJ01005235 MBW54376.1 GGFJ01005234 MBW54375.1 GGFM01008381 MBW29132.1 AEFK01088243 KQ041052 KKF31012.1 CP026254 AWP10851.1 MWRG01014020 PRD24815.1 GL194300 EFB23269.1 AEFK01102680 GBYB01013193 JAG82960.1 KQ435700 KOX80499.1 HAEA01008761 SBQ37241.1 GBYB01013192 JAG82959.1 HADY01020286 SBP58771.1 GACK01007555 JAA57479.1 HAEF01013857 SBR55016.1 CAAE01014638 CAG01317.1 HADW01017882 HADX01008529 SBP19282.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000027135
UP000245037
+ More
UP000092461 UP000028760 UP000261500 UP000261480 UP000054359 UP000007267 UP000257200 UP000261400 UP000192247 UP000007110 UP000001555 UP000265080 UP000257160 UP000186698 UP000000673 UP000286642 UP000192220 UP000261440 UP000265000 UP000075920 UP000261360 UP000264820 UP000261420 UP000007648 UP000265040 UP000246464 UP000265120 UP000264800 UP000265020 UP000261620 UP000261640 UP000053105 UP000005226 UP000009136 UP000018467 UP000002280 UP000087266
UP000092461 UP000028760 UP000261500 UP000261480 UP000054359 UP000007267 UP000257200 UP000261400 UP000192247 UP000007110 UP000001555 UP000265080 UP000257160 UP000186698 UP000000673 UP000286642 UP000192220 UP000261440 UP000265000 UP000075920 UP000261360 UP000264820 UP000261420 UP000007648 UP000265040 UP000246464 UP000265120 UP000264800 UP000265020 UP000261620 UP000261640 UP000053105 UP000005226 UP000009136 UP000018467 UP000002280 UP000087266
PRIDE
Interpro
SUPFAM
SSF53448
SSF53448
ProteinModelPortal
H9JI19
A0A2W1C0H6
A0A212ENI5
A0A2H1V461
A0A194QC64
A0A0N1INH7
+ More
A0A2W1BR41 A0A1L8E1E9 A0A1L8E1M0 A0A067QWG9 A0A2P8Y659 A0A1L8E1W0 A0A336M552 A0A1B0CF99 A0A336MN17 A0A2P6LAA8 G3MJ91 A0A1Y1KV48 A0A1Y1KUY2 A0A087YR42 A0A3B3UX26 A0A3B3YZS7 A0A1B0GIK7 A0A2L2Y294 A0A087T2X2 A0A1Y1KZV3 K7EW89 A0A3Q1EGK1 A0A3B5B1H2 A0A1V9X4N4 W4Y0Z9 B7PGF2 A0A3P8TJH5 A0A0K2U674 A0A1B6I5F0 A0A0S7I7T0 A0A3Q1BMB2 A0A1L8F8A2 K7GBA0 A0A2M4ALW2 W5JAY7 A0A087ZCU3 A0A2M4AL69 A0A2R5LKE6 A0A1B6M1H3 A0A3Q7XHS4 A0A2I4AN74 A0A147BCK9 A0A3B4CNK0 A0A224YQ19 A0A2M4BNB4 A0A3Q2PWI1 A0A147AES7 A0A182W5R3 A0A2M4BMR3 A0A2M4BMU1 A0A3B4XW44 A0A3Q2YY68 A0A3B4V7R0 A0A2M3ZKV5 G3VGE5 A0A0F8BC69 A0A3Q1HU30 A0A2U9C2L5 A0A2P6KER0 A0A3P8V1B0 A0A3Q2ZPY1 D2I3E2 A0A3Q2DPH1 A0A3B5A4J4 A0A3Q4AN34 A0A3B4YQC7 G3VBU5 A0A3Q3L7E7 A0A0C9QFA1 A0A0M9ACS9 H2TR38 A0A3Q1F029 A0A3Q1BAD7 A0A3B4U1V3 F1MGN7 A0A1A8DVG6 W5LU59 A0A3B4ZXR7 A0A0C9RIX5 A0A1A8AWB8 L7M312 A0A1A8MEQ5 Q4SDG5 K7E0Y3 A0A1A7XMH4 H2TP59 A0A1S3MWW4
A0A2W1BR41 A0A1L8E1E9 A0A1L8E1M0 A0A067QWG9 A0A2P8Y659 A0A1L8E1W0 A0A336M552 A0A1B0CF99 A0A336MN17 A0A2P6LAA8 G3MJ91 A0A1Y1KV48 A0A1Y1KUY2 A0A087YR42 A0A3B3UX26 A0A3B3YZS7 A0A1B0GIK7 A0A2L2Y294 A0A087T2X2 A0A1Y1KZV3 K7EW89 A0A3Q1EGK1 A0A3B5B1H2 A0A1V9X4N4 W4Y0Z9 B7PGF2 A0A3P8TJH5 A0A0K2U674 A0A1B6I5F0 A0A0S7I7T0 A0A3Q1BMB2 A0A1L8F8A2 K7GBA0 A0A2M4ALW2 W5JAY7 A0A087ZCU3 A0A2M4AL69 A0A2R5LKE6 A0A1B6M1H3 A0A3Q7XHS4 A0A2I4AN74 A0A147BCK9 A0A3B4CNK0 A0A224YQ19 A0A2M4BNB4 A0A3Q2PWI1 A0A147AES7 A0A182W5R3 A0A2M4BMR3 A0A2M4BMU1 A0A3B4XW44 A0A3Q2YY68 A0A3B4V7R0 A0A2M3ZKV5 G3VGE5 A0A0F8BC69 A0A3Q1HU30 A0A2U9C2L5 A0A2P6KER0 A0A3P8V1B0 A0A3Q2ZPY1 D2I3E2 A0A3Q2DPH1 A0A3B5A4J4 A0A3Q4AN34 A0A3B4YQC7 G3VBU5 A0A3Q3L7E7 A0A0C9QFA1 A0A0M9ACS9 H2TR38 A0A3Q1F029 A0A3Q1BAD7 A0A3B4U1V3 F1MGN7 A0A1A8DVG6 W5LU59 A0A3B4ZXR7 A0A0C9RIX5 A0A1A8AWB8 L7M312 A0A1A8MEQ5 Q4SDG5 K7E0Y3 A0A1A7XMH4 H2TP59 A0A1S3MWW4
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Golgi apparatus membrane
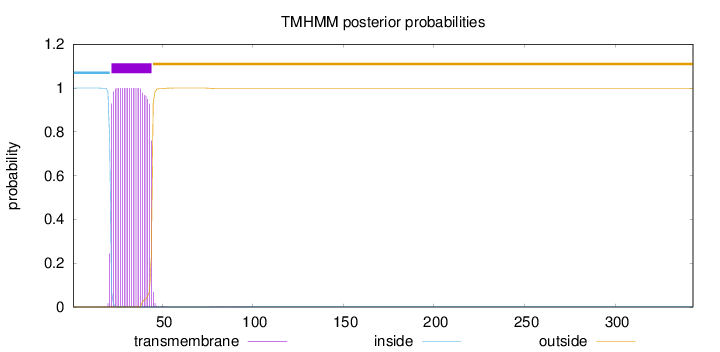
Length:
343
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.8597899999999
Exp number, first 60 AAs:
22.81959
Total prob of N-in:
0.99974
POSSIBLE N-term signal
sequence
inside
1 - 21
TMhelix
22 - 44
outside
45 - 343
Population Genetic Test Statistics
Pi
18.284772
Theta
16.584217
Tajima's D
0.096424
CLR
3.116836
CSRT
0.401129943502825
Interpretation
Uncertain
