Gene
KWMTBOMO12303 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009162
Annotation
putative_lipase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.356
Sequence
CDS
ATGGGAGAAAAGCTCGGGGCGACTTTAGCTGCTATGGTTAAAGGAGGCATGAACCCTCGAGCTATTCACCTAATCGGCCACAGCCTCGGCTCCCACATCGCAGGGTTCACCGGGAAAACCTTCACCGACCTTACGGGAAGGCGCGTCGGCAGGATTTCAGGACTGGACCCGGCCGGCCCGTGCTTCAGCAACATCGACCCCGAACTGCGGCTCAAGGCGACTGACGCTGACTTCGTGGACGTCATACACACGGACGCCGGAGTATACGGGCTCAGAGATGCCGTAGGCCACGTGGACTTTTTCCCGAACAGCGGCTCCCAGCAACCGAACTGCTTGCTGCAGACGTGCAGCCACAGCCGAGCCTGGCTCTACTACGCAGAGTCCGTGGTCAACAAGGAAGCCTTCCTCGCAGTGCCTTGTAAGGACTGGGACGCCTTTAGGAAGAATCAATGTGGCGCCGAGATAAGCATTATGGGCTACGGCACTCGGCCGAGCGCCCGCGGGGTATACTTCCTGCGCACCGCCGAGGACTCCCCCTACGGGCTCTCCCGCCGCGGGACGGTCTATGAGAACAACGAGGGCGTCATCAGGACCATCGGCTCCAAGATCCTCGGGTGA
Protein
MGEKLGATLAAMVKGGMNPRAIHLIGHSLGSHIAGFTGKTFTDLTGRRVGRISGLDPAGPCFSNIDPELRLKATDADFVDVIHTDAGVYGLRDAVGHVDFFPNSGSQQPNCLLQTCSHSRAWLYYAESVVNKEAFLAVPCKDWDAFRKNQCGAEISIMGYGTRPSARGVYFLRTAEDSPYGLSRRGTVYENNEGVIRTIGSKILG
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
D2KMR4
A0A220K8L9
A0A2W1BXY3
A0A212EJL3
A0A2W1BV63
A0A2H1W7F1
+ More
A0A2W1BWG2 A0A0L7L8R4 A0A3S2NZZ7 A0A2H1W7Q1 A0A2H1WNR8 A0A2A4J5Z0 A0A2H1W795 A0A194QC74 A0A2A4J690 A0A0N0PB11 A0A2A4IT83 A0A212FKA5 H9JFT5 A0A2H1WL84 A0A194PCA8 A0A194QNS4 A0A2A4JYV7 A0A0L7LP71 Q06VE9 A0A0L7KP71 A0A2N9QV27 A0A3S2L3L9 A0A194RQL7 A0A194PSE8 I6TFN4 A0A026WVQ0 A0A2A4JGD9 A0A3L8DFK2 A0A2P8YQG3 H9JWY8 A0A1L8DPW7 A0A2H1X3Q9 A0A1L8DPW3 A0A1L8DQ49 E0VJB4 A0A0K8VXK1 Q17N47 E2AIM4 E9IDK7 A0A034WKC4 A0A1S4EX28 A0A0K8U3C5 D6WBV6 A0A0N1PJG6 W8CB58 A0A087ZXA0 A0A2A4J1M9 A0A2W1B7Q2 K7IZY1 A0A0P4VMV4 A0A182GHZ7 A0A2H1W9L5 B4LUR2 A0A182GDZ8 A0A1L8DJ01 A0A1L8DIY9 A0A1L8DJ93 A0A1L8DIZ7 A0A0A1WYA6 A0A1B0GP74 U5EEG1 A0A087ZXA2 B3NIK0 B4PFA3 A0A1Q3FQ64 A0A1Q3FQI9 A0A1Q3FQH7 A0A1I8N4Q3 A0A146M134 A0A146LQV4 A0A2R7X607 B0X7X6 A0A224XHV1 A0A1J1IPR1 A0A146LJ26 A0A2S2R7F4 A0A3M7SXY0 A0A023G4W6 Q9VPH3 T1H9Y2 A3EXW4 A0A023G8K0 A0A1W4W6Z3 A0A158NQH7 A0A087ZXA5 A0A0U1XPW6 B0XB54 E9JDE9 A0A194Q2K6 A0A146LD99 B0X1I0 A0A1Q3FPG4 A0A146LFH2 A0A0K8WHX4
A0A2W1BWG2 A0A0L7L8R4 A0A3S2NZZ7 A0A2H1W7Q1 A0A2H1WNR8 A0A2A4J5Z0 A0A2H1W795 A0A194QC74 A0A2A4J690 A0A0N0PB11 A0A2A4IT83 A0A212FKA5 H9JFT5 A0A2H1WL84 A0A194PCA8 A0A194QNS4 A0A2A4JYV7 A0A0L7LP71 Q06VE9 A0A0L7KP71 A0A2N9QV27 A0A3S2L3L9 A0A194RQL7 A0A194PSE8 I6TFN4 A0A026WVQ0 A0A2A4JGD9 A0A3L8DFK2 A0A2P8YQG3 H9JWY8 A0A1L8DPW7 A0A2H1X3Q9 A0A1L8DPW3 A0A1L8DQ49 E0VJB4 A0A0K8VXK1 Q17N47 E2AIM4 E9IDK7 A0A034WKC4 A0A1S4EX28 A0A0K8U3C5 D6WBV6 A0A0N1PJG6 W8CB58 A0A087ZXA0 A0A2A4J1M9 A0A2W1B7Q2 K7IZY1 A0A0P4VMV4 A0A182GHZ7 A0A2H1W9L5 B4LUR2 A0A182GDZ8 A0A1L8DJ01 A0A1L8DIY9 A0A1L8DJ93 A0A1L8DIZ7 A0A0A1WYA6 A0A1B0GP74 U5EEG1 A0A087ZXA2 B3NIK0 B4PFA3 A0A1Q3FQ64 A0A1Q3FQI9 A0A1Q3FQH7 A0A1I8N4Q3 A0A146M134 A0A146LQV4 A0A2R7X607 B0X7X6 A0A224XHV1 A0A1J1IPR1 A0A146LJ26 A0A2S2R7F4 A0A3M7SXY0 A0A023G4W6 Q9VPH3 T1H9Y2 A3EXW4 A0A023G8K0 A0A1W4W6Z3 A0A158NQH7 A0A087ZXA5 A0A0U1XPW6 B0XB54 E9JDE9 A0A194Q2K6 A0A146LD99 B0X1I0 A0A1Q3FPG4 A0A146LFH2 A0A0K8WHX4
Pubmed
19121390
28630352
28756777
22118469
26227816
26354079
+ More
16876847 23171126 24508170 30249741 29403074 20566863 17510324 20798317 21282665 25348373 18362917 19820115 24495485 20075255 27129103 26483478 17994087 25830018 17550304 25315136 26823975 30375419 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 21347285
16876847 23171126 24508170 30249741 29403074 20566863 17510324 20798317 21282665 25348373 18362917 19820115 24495485 20075255 27129103 26483478 17994087 25830018 17550304 25315136 26823975 30375419 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 21347285
EMBL
BABH01035404
BABH01035405
GU230793
ADA67928.1
MF319718
ASJ26432.1
+ More
KZ149926 PZC77546.1 AGBW02014435 OWR41693.1 PZC77545.1 ODYU01006796 SOQ48977.1 PZC77547.1 JTDY01002277 KOB71750.1 RSAL01000012 RVE53468.1 SOQ48976.1 ODYU01009953 SOQ54715.1 NWSH01003103 PCG66944.1 SOQ48975.1 KQ459193 KPJ03024.1 PCG66943.1 KQ461072 KPJ09835.1 NWSH01007523 PCG62931.1 AGBW02008082 OWR54177.1 BABH01001653 ODYU01009427 SOQ53845.1 KQ459606 KPI90911.1 KQ461196 KPJ06625.1 NWSH01000413 PCG76562.1 JTDY01000429 KOB77220.1 DQ517337 ABF70649.1 JTDY01007594 KOB65092.1 KY434117 AUS94238.1 RSAL01000184 RVE44850.1 KQ459833 KPJ19782.1 KQ459601 KPI94050.1 JQ814807 AFM73623.1 KK107109 EZA59174.1 NWSH01001670 PCG70462.1 QOIP01000008 RLU19237.1 PYGN01000432 PSN46485.1 BABH01041781 GFDF01005571 JAV08513.1 ODYU01013233 SOQ59929.1 GFDF01005566 JAV08518.1 GFDF01005570 JAV08514.1 DS235221 EEB13470.1 GDHF01008697 JAI43617.1 CH477202 EAT48096.1 GL439840 EFN66713.1 GL762454 EFZ21361.1 GAKP01004764 JAC54188.1 GDHF01031494 JAI20820.1 KQ971315 EEZ97864.2 KQ460240 KPJ16392.1 GAMC01000754 JAC05802.1 NWSH01003717 PCG65891.1 KZ150304 PZC71518.1 GDKW01002938 JAI53657.1 JXUM01065049 KQ562326 KXJ76141.1 ODYU01007212 SOQ49795.1 CH940649 EDW64239.1 JXUM01056947 KQ561932 KXJ77126.1 GFDF01007648 JAV06436.1 GFDF01007645 JAV06439.1 GFDF01007647 JAV06437.1 GFDF01007646 JAV06438.1 GBXI01010636 JAD03656.1 AJVK01005768 GANO01004326 JAB55545.1 CH954178 EDV52496.1 CM000159 EDW95189.1 GFDL01005419 JAV29626.1 GFDL01005175 JAV29870.1 GFDL01005174 JAV29871.1 GDHC01006319 JAQ12310.1 GDHC01009552 JAQ09077.1 KK857139 PTY26870.1 DS232468 EDS42195.1 GFTR01004361 JAW12065.1 CVRI01000056 CRL01706.1 GDHC01010486 JAQ08143.1 GGMS01016732 MBY85935.1 REGN01000631 RNA40557.1 GBBM01006564 JAC28854.1 AE014296 AAF51579.2 ACPB03014206 EF092032 ABN12024.1 GBBM01005276 JAC30142.1 ADTU01023297 ADTU01023298 ADTU01023299 ADTU01023300 ADTU01023301 ADTU01023302 ADTU01023303 ADTU01023304 KJ018765 AIY34728.1 DS232614 EDS44107.1 GL771866 EFZ09198.1 KQ459589 KPI97640.1 GDHC01013577 JAQ05052.1 DS232260 EDS38629.1 GFDL01005544 JAV29501.1 GDHC01012863 JAQ05766.1 GDHF01001578 JAI50736.1
KZ149926 PZC77546.1 AGBW02014435 OWR41693.1 PZC77545.1 ODYU01006796 SOQ48977.1 PZC77547.1 JTDY01002277 KOB71750.1 RSAL01000012 RVE53468.1 SOQ48976.1 ODYU01009953 SOQ54715.1 NWSH01003103 PCG66944.1 SOQ48975.1 KQ459193 KPJ03024.1 PCG66943.1 KQ461072 KPJ09835.1 NWSH01007523 PCG62931.1 AGBW02008082 OWR54177.1 BABH01001653 ODYU01009427 SOQ53845.1 KQ459606 KPI90911.1 KQ461196 KPJ06625.1 NWSH01000413 PCG76562.1 JTDY01000429 KOB77220.1 DQ517337 ABF70649.1 JTDY01007594 KOB65092.1 KY434117 AUS94238.1 RSAL01000184 RVE44850.1 KQ459833 KPJ19782.1 KQ459601 KPI94050.1 JQ814807 AFM73623.1 KK107109 EZA59174.1 NWSH01001670 PCG70462.1 QOIP01000008 RLU19237.1 PYGN01000432 PSN46485.1 BABH01041781 GFDF01005571 JAV08513.1 ODYU01013233 SOQ59929.1 GFDF01005566 JAV08518.1 GFDF01005570 JAV08514.1 DS235221 EEB13470.1 GDHF01008697 JAI43617.1 CH477202 EAT48096.1 GL439840 EFN66713.1 GL762454 EFZ21361.1 GAKP01004764 JAC54188.1 GDHF01031494 JAI20820.1 KQ971315 EEZ97864.2 KQ460240 KPJ16392.1 GAMC01000754 JAC05802.1 NWSH01003717 PCG65891.1 KZ150304 PZC71518.1 GDKW01002938 JAI53657.1 JXUM01065049 KQ562326 KXJ76141.1 ODYU01007212 SOQ49795.1 CH940649 EDW64239.1 JXUM01056947 KQ561932 KXJ77126.1 GFDF01007648 JAV06436.1 GFDF01007645 JAV06439.1 GFDF01007647 JAV06437.1 GFDF01007646 JAV06438.1 GBXI01010636 JAD03656.1 AJVK01005768 GANO01004326 JAB55545.1 CH954178 EDV52496.1 CM000159 EDW95189.1 GFDL01005419 JAV29626.1 GFDL01005175 JAV29870.1 GFDL01005174 JAV29871.1 GDHC01006319 JAQ12310.1 GDHC01009552 JAQ09077.1 KK857139 PTY26870.1 DS232468 EDS42195.1 GFTR01004361 JAW12065.1 CVRI01000056 CRL01706.1 GDHC01010486 JAQ08143.1 GGMS01016732 MBY85935.1 REGN01000631 RNA40557.1 GBBM01006564 JAC28854.1 AE014296 AAF51579.2 ACPB03014206 EF092032 ABN12024.1 GBBM01005276 JAC30142.1 ADTU01023297 ADTU01023298 ADTU01023299 ADTU01023300 ADTU01023301 ADTU01023302 ADTU01023303 ADTU01023304 KJ018765 AIY34728.1 DS232614 EDS44107.1 GL771866 EFZ09198.1 KQ459589 KPI97640.1 GDHC01013577 JAQ05052.1 DS232260 EDS38629.1 GFDL01005544 JAV29501.1 GDHC01012863 JAQ05766.1 GDHF01001578 JAI50736.1
Proteomes
UP000005204
UP000007151
UP000037510
UP000283053
UP000218220
UP000053268
+ More
UP000053240 UP000001323 UP000053097 UP000279307 UP000245037 UP000009046 UP000008820 UP000000311 UP000007266 UP000005203 UP000002358 UP000069940 UP000249989 UP000008792 UP000092462 UP000008711 UP000002282 UP000095301 UP000002320 UP000183832 UP000276133 UP000000803 UP000015103 UP000192221 UP000005205
UP000053240 UP000001323 UP000053097 UP000279307 UP000245037 UP000009046 UP000008820 UP000000311 UP000007266 UP000005203 UP000002358 UP000069940 UP000249989 UP000008792 UP000092462 UP000008711 UP000002282 UP000095301 UP000002320 UP000183832 UP000276133 UP000000803 UP000015103 UP000192221 UP000005205
Pfam
PF00151 Lipase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
D2KMR4
A0A220K8L9
A0A2W1BXY3
A0A212EJL3
A0A2W1BV63
A0A2H1W7F1
+ More
A0A2W1BWG2 A0A0L7L8R4 A0A3S2NZZ7 A0A2H1W7Q1 A0A2H1WNR8 A0A2A4J5Z0 A0A2H1W795 A0A194QC74 A0A2A4J690 A0A0N0PB11 A0A2A4IT83 A0A212FKA5 H9JFT5 A0A2H1WL84 A0A194PCA8 A0A194QNS4 A0A2A4JYV7 A0A0L7LP71 Q06VE9 A0A0L7KP71 A0A2N9QV27 A0A3S2L3L9 A0A194RQL7 A0A194PSE8 I6TFN4 A0A026WVQ0 A0A2A4JGD9 A0A3L8DFK2 A0A2P8YQG3 H9JWY8 A0A1L8DPW7 A0A2H1X3Q9 A0A1L8DPW3 A0A1L8DQ49 E0VJB4 A0A0K8VXK1 Q17N47 E2AIM4 E9IDK7 A0A034WKC4 A0A1S4EX28 A0A0K8U3C5 D6WBV6 A0A0N1PJG6 W8CB58 A0A087ZXA0 A0A2A4J1M9 A0A2W1B7Q2 K7IZY1 A0A0P4VMV4 A0A182GHZ7 A0A2H1W9L5 B4LUR2 A0A182GDZ8 A0A1L8DJ01 A0A1L8DIY9 A0A1L8DJ93 A0A1L8DIZ7 A0A0A1WYA6 A0A1B0GP74 U5EEG1 A0A087ZXA2 B3NIK0 B4PFA3 A0A1Q3FQ64 A0A1Q3FQI9 A0A1Q3FQH7 A0A1I8N4Q3 A0A146M134 A0A146LQV4 A0A2R7X607 B0X7X6 A0A224XHV1 A0A1J1IPR1 A0A146LJ26 A0A2S2R7F4 A0A3M7SXY0 A0A023G4W6 Q9VPH3 T1H9Y2 A3EXW4 A0A023G8K0 A0A1W4W6Z3 A0A158NQH7 A0A087ZXA5 A0A0U1XPW6 B0XB54 E9JDE9 A0A194Q2K6 A0A146LD99 B0X1I0 A0A1Q3FPG4 A0A146LFH2 A0A0K8WHX4
A0A2W1BWG2 A0A0L7L8R4 A0A3S2NZZ7 A0A2H1W7Q1 A0A2H1WNR8 A0A2A4J5Z0 A0A2H1W795 A0A194QC74 A0A2A4J690 A0A0N0PB11 A0A2A4IT83 A0A212FKA5 H9JFT5 A0A2H1WL84 A0A194PCA8 A0A194QNS4 A0A2A4JYV7 A0A0L7LP71 Q06VE9 A0A0L7KP71 A0A2N9QV27 A0A3S2L3L9 A0A194RQL7 A0A194PSE8 I6TFN4 A0A026WVQ0 A0A2A4JGD9 A0A3L8DFK2 A0A2P8YQG3 H9JWY8 A0A1L8DPW7 A0A2H1X3Q9 A0A1L8DPW3 A0A1L8DQ49 E0VJB4 A0A0K8VXK1 Q17N47 E2AIM4 E9IDK7 A0A034WKC4 A0A1S4EX28 A0A0K8U3C5 D6WBV6 A0A0N1PJG6 W8CB58 A0A087ZXA0 A0A2A4J1M9 A0A2W1B7Q2 K7IZY1 A0A0P4VMV4 A0A182GHZ7 A0A2H1W9L5 B4LUR2 A0A182GDZ8 A0A1L8DJ01 A0A1L8DIY9 A0A1L8DJ93 A0A1L8DIZ7 A0A0A1WYA6 A0A1B0GP74 U5EEG1 A0A087ZXA2 B3NIK0 B4PFA3 A0A1Q3FQ64 A0A1Q3FQI9 A0A1Q3FQH7 A0A1I8N4Q3 A0A146M134 A0A146LQV4 A0A2R7X607 B0X7X6 A0A224XHV1 A0A1J1IPR1 A0A146LJ26 A0A2S2R7F4 A0A3M7SXY0 A0A023G4W6 Q9VPH3 T1H9Y2 A3EXW4 A0A023G8K0 A0A1W4W6Z3 A0A158NQH7 A0A087ZXA5 A0A0U1XPW6 B0XB54 E9JDE9 A0A194Q2K6 A0A146LD99 B0X1I0 A0A1Q3FPG4 A0A146LFH2 A0A0K8WHX4
PDB
1W52
E-value=3.02637e-33,
Score=351
Ontologies
GO
PANTHER
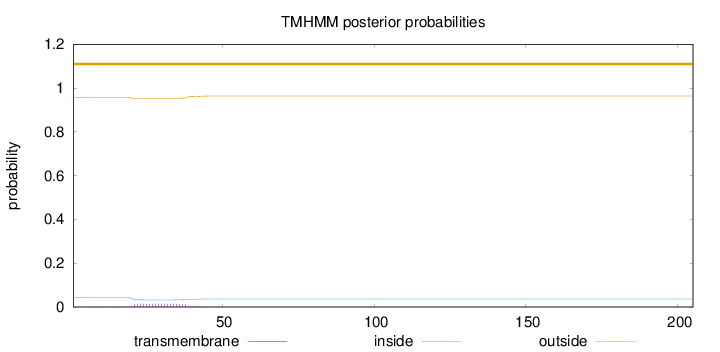
Topology
Subcellular location
Secreted
Length:
205
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.28317
Exp number, first 60 AAs:
0.2828
Total prob of N-in:
0.04283
outside
1 - 205
Population Genetic Test Statistics
Pi
14.946722
Theta
16.158273
Tajima's D
-0.446156
CLR
2.080629
CSRT
0.256287185640718
Interpretation
Uncertain
