Gene
KWMTBOMO12302
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.046
Sequence
CDS
ATGGAGTCCACGATCGATCTGTTCGCGGAATTCCTCCGTCTGAGGTACCCGACATTATCGGCCGAATTTTTAGAATTTAAGGCCAATCACGCCGCGAACTCAGAAGCGGTCTCCGTCGCGCCCGCCGCTCCCGTGTCCCCAATACTCGCGAGCAAAGCTCCCGCGTCGATCGCTGCGGTCTCGGTTCCAGCCCTGCGCTCATCCGCAGCACACGTCGCGTCCACGCGTTCATCCGCGGCCTCCATCGCGCCCGCGCCGTCCTCTGCCCCCGAGCGATTGTCCGTGGCCTCCGTCGCGACCGTCAACCCCGCGCCCTCTAAAACGCCCGCCTGCGGGTCGCCCGCGCCCTCCTCTTCATCCGACTCGGAGTCGGACATGGAGGTCGATCTTTCCCCCGCCCCCTCGACTGATGGATTCACAGTAGTCCAAAAAGGTAAGAGGCGCGCTGCGGAAGCCCGAGCTCCCGCGGCCGCCAAAGTAAGCAGAGCCGCGAACGCGTCGCGCCTCCGTCCACCGACCCCCGTTGCTCCCCCTGCCCGTGCTACACCGTCGCCGCGCCCGGTAGTACAAAAGAAAATCCAAGCCCCTCCCCCGGTAATCCTTCAAGAGAAGGCAGCCTGGGAACGAGTTTCCCTGGCCCTTAAGGCCAAAAATATTAATTTCACGAATGCCCGTAACCTCGCGAACGGCATTCAAATTAAGGTTCAAACACCCGACGACCATAGGGCCCTCTCTTCTTACCTCCGTAAGGAGCGTATAAGTTTCCATACGTATACGCTCCAGGAGGAGCGCGAACTTCGTGCCGTCATACGTGGCATCCCTAAAGAGTTGGATGCCGAACTCGTCAAGGCCGACCTTCTGGAACAAGGCCTACCAGTAAATTCAGTGCACCGCATGCACACCGGCCGCGGTAGGGAGCCATATAATATGGTTCTAGTCGCCCTCCAGCCTACCCCGAGGGAGACTCTACTTAAGCCCGCGCGCCGCGACCCTAAAATCGCGAACTATAACATGGTTAGGAACGACAGGCTCACTGCTCGTGGTGGTGGTACCGTCATCTACTACAGAAGAGCCTTGCACTGCATCCCGCTCGATCCCCCCGCGCTCGTTAACATCGAAGCATCAGTATGCCGAATCTCACTGACGGGACACGCGCCGATCGTTATCGCGTCCGTTTATCTTCCACCGGATAAGATTGTTCTAAGCAGTGATATCGAGGCGCTGCTCAGCATGGGAAGCTCTGTCATTCTGGCGGGCGACCTAAATTGTAAACACATCAGGTGGAATTCTCACACCACAACCCCTAATGGCAGGCGGCTCGACGCGCTAGTTGATGATCTCGCCTTCGATATCATCGCTCCGCTAACCCCGACTCACTACCCGCTAAATATCGCGCATCGCCCGGATATACTCGACATAGCGTTATTAAAAAACGTAGCTCTGCGCTTACACTCGATCGAAGTAGTTTCAGAGTTAGATTCAGACCACCGTCCCGTCGTTATGAAACTCGGTCGCGCTCCCGATTCCGTTCCCGTCACGAGGACTGTGGTGGATTGGCACACGCTGGGCATCAGCCTGGCTGAATCTGATCCACCATCGCTCCCGTTTAGCCCGGACTCTACCCCGTCTCTTCAGGATACCGCTGAAGCCATAGACATCCTAACGTCACACATTACCTCGACATTAGATAGGTCATCGAAGCAAGTTGTAGCGGAGGATTTCCTTCACCGCTTTGAATTGCCCGACGATATTAGGGAACTCCTTAGAGCTAAGAACGCTTCGATCCGCGCCTACGATAGGTATCCTACCATGGAAAATCGTATTCGAATGCGTGCCCTACAACGCGACGTAAAGTCTCGCATCGCCGAAATCCGAGATGCTAGGTGGTCTGATTTCTTAGAAGGACTCGCGCCCTCTCAAAGGTCTTACTACCGTTTAGCTCGTACTCTCAAATCGGATACGGTTGTAACTATGCCCCCCCTCGTAGGCCCCTCAGGTCGACTCGCGGCGTTTGATGATGACGAAAAAGCAGAGCTGCTGGCCGATACATTGCAAACCCAGTGCACGCCCAGCATTCAATCCGCGGACCCTGTTCATGTAGAATTAGTAGACAGTGAGGTAGAACGCAGAGCCTCCTTGCCACCCTCGGATGCGTTACCACCCGTCACTCCAATGGAAGTTAAAGATCTGATCAAAGACCTACGTCCTCGCAAGGCTCCCGGTTCCGACGGTATATCTAACCGCGTTATTAAACTTCTACCCATCCAACTCATCGTGATGTTGGCATCTATTTTCAATGCCGCTATGGCGAACTGTATCTTTCCCGCGGTGTGGAAAGAAGCAGACGTTATCGGCATACATAAACCCGGTAAACCAAAAAATCATCCGACGAGTTACCGCCCGATTAGCCTCCTCATGTCTCTGGGCAAACTGTATGAGCGTTTGCTCTACAAACGCCTCAGAGACTTCGTCTCATCAAAGGGCATTCTCATCGATGAACAATTCGGATTCCGTGCAAATCACTCATGCGTACAACAGGTGCACCGCCTCACGGAGCACATTCTTGTAGGACTTAACCGACCAAAACCGATCTATACGGGAGCCCTCTTCTTCGACGTCGCAAAAGCGTTCGACAAAGTCTGGCACAACGGTTTGATTTTCAAACTTTTCAACATGGGCGTACCGGATAGTCTCGTGCTCATCATACGAGACTTCTTGTCTAACCGCTCTTTTCGATATCGAGTAGAGGGAACCCGCTCCTCCCCGCGACCACTCACGGCTGGAGTCCCGCAAGGCTCCGTTCTCTCTCCGCTCCTATTTAGTTTATTTATTAACGATATTCCCCGGTCGCCGCCGACTCAGCTAGCCTTATTCGCTGACGACACAACCGTTTACTACTCAAGTAGAAACAAGTCCCTAATCGCGAGTAAGCTCCAGAGTGCAGCGTTAACCCTCGGACAGTGGTTCCGAAAATGGCGCATAGACATCAACCCAGCGAAAAGCACAGCAGTGTTATTTCAAAGGGGAAACTCACCGCTTATTTCCTCCCGCATTAGGAGGAGAAATATCACACCCCCGATCACCCTCTTCGGCCAACCTATACCCTGGGCTAGGAAGGTCAAGTACCTGGGAGTCACCCTGGATGCATCCATGACATTCCGCCCGCATATAAAAACAGTCCGCGATCGTGCCGCATTTATTCTCGGCAGACTCTATCCCATGATCTGTAAGCGGAGTAAAATGTCCCTTCGGAACAAGGTGACACTTTACAAAACTTGCATAAGGCCCGTCATGACTTACGCGAGTGTGGTGTTCGCTCACGCGGCCCGCACACACATAGACACCCTCCAATCCCTACAATCCCGCTTTTGCAGGTTAGCCGTCGGGGCTCCGTGGTTCGTGAGGAACGTCGACCTACACGACGACCTGGGCCTCGAATCGATCCGCAAACACATGAAGTCAGTGTCGGAACGTTACTTCGATAAGGCTATGCGTCATGATAATCGCCTTATCGTTGCTGCCGCTGACTACTCCCCGAATCCTGATCACGCAGGAGCAAGTCACCGTCGTCGCCCTAGACACGTCCTTACGGATCCATCAGATCCAATAACTCTTGCATTAGATACCTTCAGCTCTAACACTAGGAGCAGGCTGAGGAACCCCGGTAACCGTACGCGTCGAACTCGACAAAGAGGTCGACGTGCAGCCTAA
Protein
MESTIDLFAEFLRLRYPTLSAEFLEFKANHAANSEAVSVAPAAPVSPILASKAPASIAAVSVPALRSSAAHVASTRSSAASIAPAPSSAPERLSVASVATVNPAPSKTPACGSPAPSSSSDSESDMEVDLSPAPSTDGFTVVQKGKRRAAEARAPAAAKVSRAANASRLRPPTPVAPPARATPSPRPVVQKKIQAPPPVILQEKAAWERVSLALKAKNINFTNARNLANGIQIKVQTPDDHRALSSYLRKERISFHTYTLQEERELRAVIRGIPKELDAELVKADLLEQGLPVNSVHRMHTGRGREPYNMVLVALQPTPRETLLKPARRDPKIANYNMVRNDRLTARGGGTVIYYRRALHCIPLDPPALVNIEASVCRISLTGHAPIVIASVYLPPDKIVLSSDIEALLSMGSSVILAGDLNCKHIRWNSHTTTPNGRRLDALVDDLAFDIIAPLTPTHYPLNIAHRPDILDIALLKNVALRLHSIEVVSELDSDHRPVVMKLGRAPDSVPVTRTVVDWHTLGISLAESDPPSLPFSPDSTPSLQDTAEAIDILTSHITSTLDRSSKQVVAEDFLHRFELPDDIRELLRAKNASIRAYDRYPTMENRIRMRALQRDVKSRIAEIRDARWSDFLEGLAPSQRSYYRLARTLKSDTVVTMPPLVGPSGRLAAFDDDEKAELLADTLQTQCTPSIQSADPVHVELVDSEVERRASLPPSDALPPVTPMEVKDLIKDLRPRKAPGSDGISNRVIKLLPIQLIVMLASIFNAAMANCIFPAVWKEADVIGIHKPGKPKNHPTSYRPISLLMSLGKLYERLLYKRLRDFVSSKGILIDEQFGFRANHSCVQQVHRLTEHILVGLNRPKPIYTGALFFDVAKAFDKVWHNGLIFKLFNMGVPDSLVLIIRDFLSNRSFRYRVEGTRSSPRPLTAGVPQGSVLSPLLFSLFINDIPRSPPTQLALFADDTTVYYSSRNKSLIASKLQSAALTLGQWFRKWRIDINPAKSTAVLFQRGNSPLISSRIRRRNITPPITLFGQPIPWARKVKYLGVTLDASMTFRPHIKTVRDRAAFILGRLYPMICKRSKMSLRNKVTLYKTCIRPVMTYASVVFAHAARTHIDTLQSLQSRFCRLAVGAPWFVRNVDLHDDLGLESIRKHMKSVSERYFDKAMRHDNRLIVAAADYSPNPDHAGASHRRRPRHVLTDPSDPITLALDTFSSNTRSRLRNPGNRTRRTRQRGRRAA
Summary
Uniprot
EMBL
AB018558
BAA76304.1
U07847
AAA17752.1
AB055391
BAB21761.1
+ More
KQ459265 KPJ02064.1 AF081103 AAC72921.1 GEZM01008457 JAV94772.1 GALX01005299 JAB63167.1 KQ971729 KYB24751.1 GALX01005359 JAB63107.1 ABLF02042518 ABLF02057746 ABLF02034467 NEVH01011985 PNF31054.1 NEVH01021205 PNF19912.1 NEVH01027074 PNF13841.1 NEVH01009134 PNF33528.1 GGMS01002992 MBY72195.1 NEVH01006736 PNF36716.1 KK119261 KFM75202.1 NEVH01017470 PNF24199.1 ABLF02023911 GFTR01008251 JAW08175.1 GBGD01000415 JAC88474.1 GGFJ01001822 MBW50963.1 GGMR01012443 MBY25062.1 GGMR01006408 MBY19027.1 GGFK01007981 MBW41302.1
KQ459265 KPJ02064.1 AF081103 AAC72921.1 GEZM01008457 JAV94772.1 GALX01005299 JAB63167.1 KQ971729 KYB24751.1 GALX01005359 JAB63107.1 ABLF02042518 ABLF02057746 ABLF02034467 NEVH01011985 PNF31054.1 NEVH01021205 PNF19912.1 NEVH01027074 PNF13841.1 NEVH01009134 PNF33528.1 GGMS01002992 MBY72195.1 NEVH01006736 PNF36716.1 KK119261 KFM75202.1 NEVH01017470 PNF24199.1 ABLF02023911 GFTR01008251 JAW08175.1 GBGD01000415 JAC88474.1 GGFJ01001822 MBW50963.1 GGMR01012443 MBY25062.1 GGMR01006408 MBY19027.1 GGFK01007981 MBW41302.1
Proteomes
Pfam
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
PDB
6AR3
E-value=0.0441353,
Score=91
Ontologies
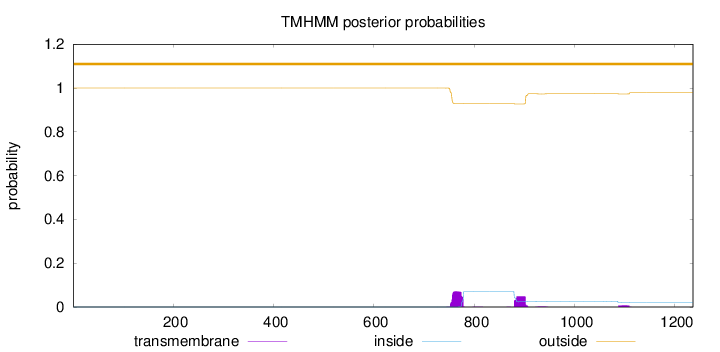
Topology
Length:
1236
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.87756
Exp number, first 60 AAs:
0.00116
Total prob of N-in:
0.00011
outside
1 - 1236
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
27.100494
CSRT
0
Interpretation
Uncertain
