Gene
KWMTBOMO12301
Annotation
PREDICTED:_protein_4.1-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.615
Sequence
CDS
ATGAGCTACAACAAGCGCTCGTTTACCCTGAAGCTGCGGGCCAGCGAGTTCGATGAGTTCGAGACGGACGTGAGCTTCAAGCTGCCCTCGACGAGAGCTTGCAAGAAGCTGTGGAAGTGCAGCGTGGAGCATCACATATTTTTTAGACGAGAGGCTCCGGTGTCTGTCCCGGGTCGGCACGGAGCCTGGGGCTCTCCGTCCTGCCGCCGCACGCTCCGCCAGCTGCGGGCGCGCCCACCACACCTGACCTCCTGA
Protein
MSYNKRSFTLKLRASEFDEFETDVSFKLPSTRACKKLWKCSVEHHIFFRREAPVSVPGRHGAWGSPSCRRTLRQLRARPPHLTS
Summary
Uniprot
A0A2A4J0H2
A0A2H1VWK5
A0A0N1IFE0
A0A194QDI0
A0A2W1BR31
A0A212EJP0
+ More
A0A3Q1AZV1 A0A3Q1FU61 A0A3Q1CYY0 A0A3Q1FWB0 A0A1A8QDM7 A0A3Q1GVE9 A0A3P8UCE1 A0A3Q1AZV9 A0A1A7XFM6 A0A3Q1FUF4 A0A3Q3BNI0 A0A1A8KQT2 A0A1A7WGJ3 A0A3Q3BNJ3 A0A3Q3BBV2 A0A1A8MR75 A0A3Q3FHL2 A0A3P8U774 A0A087XC40 A0A0S7FI64 A0A3Q3MFD5 A0A1A7X9H2 M3ZF02 A0A3P9DE65 A0A3P9DER4 A0A3P9DET3 A0A3B5QJ86 A0A3Q1CE99 A0A3Q1B9W1 A0A3Q3BUN3 A0A3Q2WU39 A0A3Q2V7L6 A0A146ZZW1 A0A3Q2V7M3 A0A146W3I4 G3PWM6 A0A1A8BFX1 A0A3Q3QZ59 A0A0S7FMI2 A0A3Q3X9M5 A0A1A7WC69 A0A0S7FL82 A0A1A7ZSP6 A0A3P9P1M3 A0A3P9P1F3 A0A3B3TP05 A0A1A8INH6 A0A2I4D336 A0A1A7ZVA3 A0A3Q4BYX4 A0A3P9P1I4 A0A3Q3LJF7 A0A2I4D316 A0A2I4D314 A0A3Q3L6G9 A0A3P9P1C3 A0A2I4D323 A0A2I4D327 A0A2I4D328 A0A3Q3LHW3 A0A3B4U2H2 A0A2I4D341 A0A2I4D332 A0A2I4D333 A0A3P8U7A9 A0A0S7FGK0 A0A3B3XS50 A0A2I4D329 A0A0S7FED0 A0A1A8JLG7 A0A3B3XSL8 A0A2I4D318 A0A3Q3LHT5 A0A1A8QW15 A0A1A8SGU9 A0A3B3V5X4 A0A1A8CX57 A0A2I4D324 A0A1A7Y674 A0A1A8JJM0 A0A1A7XDM2 A0A3Q2CCZ0 A0A1A8QQJ2
A0A3Q1AZV1 A0A3Q1FU61 A0A3Q1CYY0 A0A3Q1FWB0 A0A1A8QDM7 A0A3Q1GVE9 A0A3P8UCE1 A0A3Q1AZV9 A0A1A7XFM6 A0A3Q1FUF4 A0A3Q3BNI0 A0A1A8KQT2 A0A1A7WGJ3 A0A3Q3BNJ3 A0A3Q3BBV2 A0A1A8MR75 A0A3Q3FHL2 A0A3P8U774 A0A087XC40 A0A0S7FI64 A0A3Q3MFD5 A0A1A7X9H2 M3ZF02 A0A3P9DE65 A0A3P9DER4 A0A3P9DET3 A0A3B5QJ86 A0A3Q1CE99 A0A3Q1B9W1 A0A3Q3BUN3 A0A3Q2WU39 A0A3Q2V7L6 A0A146ZZW1 A0A3Q2V7M3 A0A146W3I4 G3PWM6 A0A1A8BFX1 A0A3Q3QZ59 A0A0S7FMI2 A0A3Q3X9M5 A0A1A7WC69 A0A0S7FL82 A0A1A7ZSP6 A0A3P9P1M3 A0A3P9P1F3 A0A3B3TP05 A0A1A8INH6 A0A2I4D336 A0A1A7ZVA3 A0A3Q4BYX4 A0A3P9P1I4 A0A3Q3LJF7 A0A2I4D316 A0A2I4D314 A0A3Q3L6G9 A0A3P9P1C3 A0A2I4D323 A0A2I4D327 A0A2I4D328 A0A3Q3LHW3 A0A3B4U2H2 A0A2I4D341 A0A2I4D332 A0A2I4D333 A0A3P8U7A9 A0A0S7FGK0 A0A3B3XS50 A0A2I4D329 A0A0S7FED0 A0A1A8JLG7 A0A3B3XSL8 A0A2I4D318 A0A3Q3LHT5 A0A1A8QW15 A0A1A8SGU9 A0A3B3V5X4 A0A1A8CX57 A0A2I4D324 A0A1A7Y674 A0A1A8JJM0 A0A1A7XDM2 A0A3Q2CCZ0 A0A1A8QQJ2
EMBL
NWSH01004116
PCG65481.1
ODYU01004882
SOQ45210.1
KQ461072
KPJ09834.1
+ More
KQ459193 KPJ03025.1 KZ149926 PZC77562.1 AGBW02014435 OWR41694.1 HAEI01005029 SBR91462.1 HADW01015250 SBP16650.1 HAEE01013959 SBR34009.1 HADW01003697 SBP05097.1 HAEF01017805 SBR58964.1 AYCK01010049 GBYX01466100 JAO15515.1 HADW01013183 SBP14583.1 GCES01014818 JAR71505.1 GCES01061735 JAR24588.1 HADZ01001857 SBP65798.1 GBYX01466102 JAO15513.1 HADW01002207 SBP03607.1 GBYX01466098 JAO15517.1 HADY01007309 SBP45794.1 HAED01012425 SBQ98819.1 HADY01008322 SBP46807.1 GBYX01466103 GBYX01466099 JAO15512.1 GBYX01466096 JAO15519.1 HAEE01000901 SBR20917.1 HAEH01013564 SBR97717.1 HAEI01014978 SBS17447.1 HADZ01019474 SBP83415.1 HADX01003566 SBP25798.1 HAEE01000285 SBR20301.1 HADW01014555 SBP15955.1 HAEH01012735 SBR95548.1
KQ459193 KPJ03025.1 KZ149926 PZC77562.1 AGBW02014435 OWR41694.1 HAEI01005029 SBR91462.1 HADW01015250 SBP16650.1 HAEE01013959 SBR34009.1 HADW01003697 SBP05097.1 HAEF01017805 SBR58964.1 AYCK01010049 GBYX01466100 JAO15515.1 HADW01013183 SBP14583.1 GCES01014818 JAR71505.1 GCES01061735 JAR24588.1 HADZ01001857 SBP65798.1 GBYX01466102 JAO15513.1 HADW01002207 SBP03607.1 GBYX01466098 JAO15517.1 HADY01007309 SBP45794.1 HAED01012425 SBQ98819.1 HADY01008322 SBP46807.1 GBYX01466103 GBYX01466099 JAO15512.1 GBYX01466096 JAO15519.1 HAEE01000901 SBR20917.1 HAEH01013564 SBR97717.1 HAEI01014978 SBS17447.1 HADZ01019474 SBP83415.1 HADX01003566 SBP25798.1 HAEE01000285 SBR20301.1 HADW01014555 SBP15955.1 HAEH01012735 SBR95548.1
Proteomes
Interpro
IPR019747
FERM_CS
+ More
IPR018979 FERM_N
IPR011993 PH-like_dom_sf
IPR019748 FERM_central
IPR018980 FERM_PH-like_C
IPR029071 Ubiquitin-like_domsf
IPR000299 FERM_domain
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR019749 Band_41_domain
IPR035963 FERM_2
IPR000798 Ez/rad/moesin-like
IPR021187 Band_41_protein_chordates
IPR014847 FERM-adjacent
IPR030691 Band4.1-L3
IPR008379 Band_4.1_C
IPR007477 SAB_dom
IPR018979 FERM_N
IPR011993 PH-like_dom_sf
IPR019748 FERM_central
IPR018980 FERM_PH-like_C
IPR029071 Ubiquitin-like_domsf
IPR000299 FERM_domain
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR019749 Band_41_domain
IPR035963 FERM_2
IPR000798 Ez/rad/moesin-like
IPR021187 Band_41_protein_chordates
IPR014847 FERM-adjacent
IPR030691 Band4.1-L3
IPR008379 Band_4.1_C
IPR007477 SAB_dom
Gene 3D
ProteinModelPortal
A0A2A4J0H2
A0A2H1VWK5
A0A0N1IFE0
A0A194QDI0
A0A2W1BR31
A0A212EJP0
+ More
A0A3Q1AZV1 A0A3Q1FU61 A0A3Q1CYY0 A0A3Q1FWB0 A0A1A8QDM7 A0A3Q1GVE9 A0A3P8UCE1 A0A3Q1AZV9 A0A1A7XFM6 A0A3Q1FUF4 A0A3Q3BNI0 A0A1A8KQT2 A0A1A7WGJ3 A0A3Q3BNJ3 A0A3Q3BBV2 A0A1A8MR75 A0A3Q3FHL2 A0A3P8U774 A0A087XC40 A0A0S7FI64 A0A3Q3MFD5 A0A1A7X9H2 M3ZF02 A0A3P9DE65 A0A3P9DER4 A0A3P9DET3 A0A3B5QJ86 A0A3Q1CE99 A0A3Q1B9W1 A0A3Q3BUN3 A0A3Q2WU39 A0A3Q2V7L6 A0A146ZZW1 A0A3Q2V7M3 A0A146W3I4 G3PWM6 A0A1A8BFX1 A0A3Q3QZ59 A0A0S7FMI2 A0A3Q3X9M5 A0A1A7WC69 A0A0S7FL82 A0A1A7ZSP6 A0A3P9P1M3 A0A3P9P1F3 A0A3B3TP05 A0A1A8INH6 A0A2I4D336 A0A1A7ZVA3 A0A3Q4BYX4 A0A3P9P1I4 A0A3Q3LJF7 A0A2I4D316 A0A2I4D314 A0A3Q3L6G9 A0A3P9P1C3 A0A2I4D323 A0A2I4D327 A0A2I4D328 A0A3Q3LHW3 A0A3B4U2H2 A0A2I4D341 A0A2I4D332 A0A2I4D333 A0A3P8U7A9 A0A0S7FGK0 A0A3B3XS50 A0A2I4D329 A0A0S7FED0 A0A1A8JLG7 A0A3B3XSL8 A0A2I4D318 A0A3Q3LHT5 A0A1A8QW15 A0A1A8SGU9 A0A3B3V5X4 A0A1A8CX57 A0A2I4D324 A0A1A7Y674 A0A1A8JJM0 A0A1A7XDM2 A0A3Q2CCZ0 A0A1A8QQJ2
A0A3Q1AZV1 A0A3Q1FU61 A0A3Q1CYY0 A0A3Q1FWB0 A0A1A8QDM7 A0A3Q1GVE9 A0A3P8UCE1 A0A3Q1AZV9 A0A1A7XFM6 A0A3Q1FUF4 A0A3Q3BNI0 A0A1A8KQT2 A0A1A7WGJ3 A0A3Q3BNJ3 A0A3Q3BBV2 A0A1A8MR75 A0A3Q3FHL2 A0A3P8U774 A0A087XC40 A0A0S7FI64 A0A3Q3MFD5 A0A1A7X9H2 M3ZF02 A0A3P9DE65 A0A3P9DER4 A0A3P9DET3 A0A3B5QJ86 A0A3Q1CE99 A0A3Q1B9W1 A0A3Q3BUN3 A0A3Q2WU39 A0A3Q2V7L6 A0A146ZZW1 A0A3Q2V7M3 A0A146W3I4 G3PWM6 A0A1A8BFX1 A0A3Q3QZ59 A0A0S7FMI2 A0A3Q3X9M5 A0A1A7WC69 A0A0S7FL82 A0A1A7ZSP6 A0A3P9P1M3 A0A3P9P1F3 A0A3B3TP05 A0A1A8INH6 A0A2I4D336 A0A1A7ZVA3 A0A3Q4BYX4 A0A3P9P1I4 A0A3Q3LJF7 A0A2I4D316 A0A2I4D314 A0A3Q3L6G9 A0A3P9P1C3 A0A2I4D323 A0A2I4D327 A0A2I4D328 A0A3Q3LHW3 A0A3B4U2H2 A0A2I4D341 A0A2I4D332 A0A2I4D333 A0A3P8U7A9 A0A0S7FGK0 A0A3B3XS50 A0A2I4D329 A0A0S7FED0 A0A1A8JLG7 A0A3B3XSL8 A0A2I4D318 A0A3Q3LHT5 A0A1A8QW15 A0A1A8SGU9 A0A3B3V5X4 A0A1A8CX57 A0A2I4D324 A0A1A7Y674 A0A1A8JJM0 A0A1A7XDM2 A0A3Q2CCZ0 A0A1A8QQJ2
PDB
3BIN
E-value=3.94812e-11,
Score=157
Ontologies
GO
PANTHER
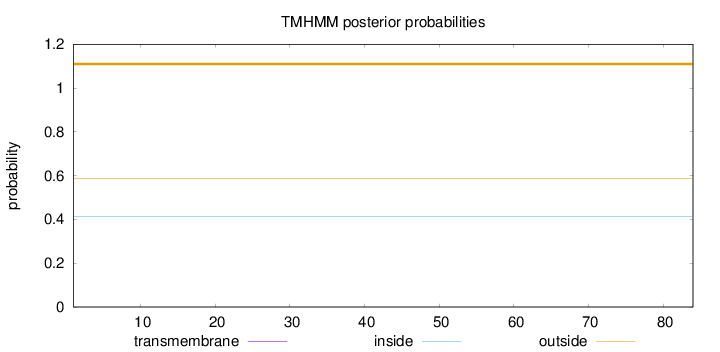
Topology
Length:
84
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.41176
outside
1 - 84
Population Genetic Test Statistics
Pi
16.256893
Theta
19.990755
Tajima's D
-0.602316
CLR
1.843424
CSRT
0.218139093045348
Interpretation
Uncertain
