Pre Gene Modal
BGIBMGA009122
Annotation
PREDICTED:_nucleolar_protein_9_isoform_X1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.913 Nuclear Reliability : 2.124
Sequence
CDS
ATGAGCGAAGAAACACAAGTTAATGATGAAAATAAAAAGAGAACGCGTAAGAAACGTAAGAATTTTCTTTCGAATGCCAAGAAATATGCCAAGAAAGGTCAAATGGGCAGGGGAACAAAAATGCCCGAGGAACTATACCAATATTTCGTTGGTATACTCGACGCTATAAAAAAGGGAATTGAAAATCAAGAGGAAAGAAATGTCCTTGTAAACAATGTACTTGAGAGAACGAAGGATGAAGAAATTAATATAATTGGGAACCAGCTTGGATGTCGGGTAATCGAGCTTCTACTACCTTATGCATCACCAGCAGACCTAGAACGGTTTATCAACGCCGTGACCCCTGAACTACGAAGACTCTGCTCGGATAATTTCTCTAGTCACGTCATAGAAACATTACTTAGAGTATCGTGTACACGTGCAACTGAGCATTTGCAGAATCTTGATGATTCAGAGGATTCTGATGAAATTCCTAAGAAGAAAGTTAAATTGGAGATTGTCTCTAAGTACAGTGATGAGCATGTAAAAACTTGTTCTGAATTTACATTGAAGCTATGTAAATATGCACTAAATAATCTAGAGGATTTTGTTTGGGATAATTATGCTAATCATATATTGAGAAGTGCTTTGAAGTGTCTTAGTGGTATTGAGCTACTGCCTGGTGAGAAGCCAAAGGTTAATCTCTTCAAAGAAACTGTTGGAGGAAAGAAAGGTATACCACCTCACATATCAAAGATGGCCTACAAAAATGTTCCTGATGAATACAAAGGTATAGTTAGAGAGTTTGCTGAGAGACTCTCCTCATGGCCACAATTTAAGGATATGGGTTACCAAAATCTGACTTCAGCATTGTTACAGGTTTTACTCTATGCAATAAAGAATGTGGATAAGAACTTAACCAAGAACATTTTAAAGAAACTTTTAAATGAAAGCTTTGCACCAGACGATTGGATCCCTAATGCTGACGATGAAAAAAACAAAGATTCTGATGAGAAAAAGGCGACTCTGAATGAGGATGGTGATGAAAAGGTGGACCTTGCAATTGAATCTGGGGGTGTGGTGCAAAGGAACTTACCACCAGTATTTGATTCAGAGTCAGCTGTAAGGTTGTTAGAAGCTGCATTGTTTGTGGCTAAAAAGAAGATGTATACCCAGATATACGCAAAGTGTTTCATCAACCGCCTCGGCCAGCTCGCCACCATGCCCATGTTGAACTTCACGGTCCAGAGACTGATCGACAACTGTGATATCAAAGAGGAGTTTGAACCAATGTTTGATGAGCTGTCGGACAAGTTCGACGCGCTGTTGGCCTGCGGGAACACCGGGGTGCTGGTGGCCCTCGCCAAAGCCTGCCTCAGGATCAAAGTGAAACAAATGCAATTTGTTAATACACTAGAAGCGGCATTGAAGTGTACAGAGGAGGGTAGGCAGAAATACTTCCCGATAGCTTGTCTCCGGCTGGTGCCGATTGACAGGATCGAGCTGGACAAGCTGGACCAGGAGTACTTCATTAACGTCCACGGATCGGTGATACTACAGACCATTCTAAATTTTCAGCGTCCGGGGCGAGCGACGGGCGCGCTGCTGGAGTTGCCGGCGGAGCAGCTGCTGGTGGTGCTGCGCGACGCGAAGGGCTGCCACGTGGCGGACGCTCTCTGCACCGGACCCTTCGTCGGCGTCAAGGCCAGGGACAAACTCATTTGGAAGCTTAAAGGCTGCTACCAGCAGCTGGCGCTGTCACAGCACGGCTCAAGGGCCTTCGAGCAAGTCTTCAGCGTCGCCTCCATGGAACAGAAACTCAAGATTATGACGGAAATCTCCGAGAAGAGTAACCTACTAAATGGCTCCGCATACGGCCGCCTGATCGCCGACAAGCTCCACGTCGCCGCCTTCAAGACCTCGCAGAAGAAATGGGAGGAAGCCTGGAGGAAACGTTCTTGCGATATAAATAGCTGA
Protein
MSEETQVNDENKKRTRKKRKNFLSNAKKYAKKGQMGRGTKMPEELYQYFVGILDAIKKGIENQEERNVLVNNVLERTKDEEINIIGNQLGCRVIELLLPYASPADLERFINAVTPELRRLCSDNFSSHVIETLLRVSCTRATEHLQNLDDSEDSDEIPKKKVKLEIVSKYSDEHVKTCSEFTLKLCKYALNNLEDFVWDNYANHILRSALKCLSGIELLPGEKPKVNLFKETVGGKKGIPPHISKMAYKNVPDEYKGIVREFAERLSSWPQFKDMGYQNLTSALLQVLLYAIKNVDKNLTKNILKKLLNESFAPDDWIPNADDEKNKDSDEKKATLNEDGDEKVDLAIESGGVVQRNLPPVFDSESAVRLLEAALFVAKKKMYTQIYAKCFINRLGQLATMPMLNFTVQRLIDNCDIKEEFEPMFDELSDKFDALLACGNTGVLVALAKACLRIKVKQMQFVNTLEAALKCTEEGRQKYFPIACLRLVPIDRIELDKLDQEYFINVHGSVILQTILNFQRPGRATGALLELPAEQLLVVLRDAKGCHVADALCTGPFVGVKARDKLIWKLKGCYQQLALSQHGSRAFEQVFSVASMEQKLKIMTEISEKSNLLNGSAYGRLIADKLHVAAFKTSQKKWEEAWRKRSCDINS
Summary
Uniprot
H9JHX4
A0A2W1BT90
A0A2A4JPI7
A0A194QHU4
A0A212EJN9
A0A0L7KRR8
+ More
W8BS29 A0A1I8PG43 B4P1P0 D6WS25 Q7JX95 A0A1A9XFG2 B4QE98 B4LMA6 B4KND5 B3N9Z7 A0A1Y1K169 A0A1I8M7W0 A0A1W4VD87 B4J6P0 A0A067R7I5 A0A034W0I0 B3MDQ0 A0A1B0AGG7 A0A1A9WR41 A0A3B0JD43 Q292S4 B4GCQ0 A0A0M3QUK8 A0A1B0G1H5 U4UCI4 A0A0L0BVZ0 A0A1W4WXZ4 N6TB60 A0A1B0BAP1 B4MR70 A0A336MV57 A0A154PB07 A0A0C9RFG5 Q16SD4 A0A2M4ASG8 K7IQP3 A0A182GIV0 A0A0L7RE32 A0A2A3EDV4 E2AQX1 A0A182MCU6 A0A084WBG4 W5JLY3 A0A182FTC1 A0A087ZYY8 E2BTB4 A0A151JSB8 A0A1Q3EYA6 A0A182WF41 A0A158NZA4 A0A195B1K1 A0A1Q3EYA3 A0A195CWL0 A0A195FTG4 E9IYS2 A0A151X374 F4WFQ5 B0WFZ1 A0A0J7K3J0 A0A182QVR7 A0A182MXY0 A0A2S2QXI9 A0A182P9E5 A0A2H8TF14 A0A182V1A3 A0A182LPK0 A0A182J894 A0A182HNM3 A0A182TTH8 A0A182XCC0 A0A2S2PEG3 A0A1Q3EY22 A0A1B6DSA6 A0A182K091 A0A026WQG8 J9JWR3 E9HMA4 A0A182RFC1 A0A162NY84 A0A0P6AGR6 A0A0P5EAK8 A0A0P6HAV8 E0W282 A0A0P5FYU1 A0A0P5ZDD5 A0A182YDC5 A0A182S8L5 A0A2R7W8U2 Q7Q318
W8BS29 A0A1I8PG43 B4P1P0 D6WS25 Q7JX95 A0A1A9XFG2 B4QE98 B4LMA6 B4KND5 B3N9Z7 A0A1Y1K169 A0A1I8M7W0 A0A1W4VD87 B4J6P0 A0A067R7I5 A0A034W0I0 B3MDQ0 A0A1B0AGG7 A0A1A9WR41 A0A3B0JD43 Q292S4 B4GCQ0 A0A0M3QUK8 A0A1B0G1H5 U4UCI4 A0A0L0BVZ0 A0A1W4WXZ4 N6TB60 A0A1B0BAP1 B4MR70 A0A336MV57 A0A154PB07 A0A0C9RFG5 Q16SD4 A0A2M4ASG8 K7IQP3 A0A182GIV0 A0A0L7RE32 A0A2A3EDV4 E2AQX1 A0A182MCU6 A0A084WBG4 W5JLY3 A0A182FTC1 A0A087ZYY8 E2BTB4 A0A151JSB8 A0A1Q3EYA6 A0A182WF41 A0A158NZA4 A0A195B1K1 A0A1Q3EYA3 A0A195CWL0 A0A195FTG4 E9IYS2 A0A151X374 F4WFQ5 B0WFZ1 A0A0J7K3J0 A0A182QVR7 A0A182MXY0 A0A2S2QXI9 A0A182P9E5 A0A2H8TF14 A0A182V1A3 A0A182LPK0 A0A182J894 A0A182HNM3 A0A182TTH8 A0A182XCC0 A0A2S2PEG3 A0A1Q3EY22 A0A1B6DSA6 A0A182K091 A0A026WQG8 J9JWR3 E9HMA4 A0A182RFC1 A0A162NY84 A0A0P6AGR6 A0A0P5EAK8 A0A0P6HAV8 E0W282 A0A0P5FYU1 A0A0P5ZDD5 A0A182YDC5 A0A182S8L5 A0A2R7W8U2 Q7Q318
Pubmed
19121390
28756777
26354079
22118469
26227816
24495485
+ More
17994087 17550304 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 28004739 25315136 24845553 25348373 15632085 23537049 26108605 17510324 20075255 26483478 20798317 24438588 20920257 23761445 21347285 21282665 21719571 20966253 24508170 30249741 21292972 20566863 25244985 12364791
17994087 17550304 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 28004739 25315136 24845553 25348373 15632085 23537049 26108605 17510324 20075255 26483478 20798317 24438588 20920257 23761445 21347285 21282665 21719571 20966253 24508170 30249741 21292972 20566863 25244985 12364791
EMBL
BABH01035387
BABH01035388
BABH01035389
BABH01035390
BABH01035391
KZ149926
+ More
PZC77561.1 NWSH01000828 PCG73967.1 KQ459193 KPJ03026.1 AGBW02014435 OWR41695.1 JTDY01006457 KOB65997.1 GAMC01002380 JAC04176.1 CM000157 EDW89176.1 KQ971354 EFA07079.1 AE013599 AY094923 AAF59259.1 AAM11276.1 CM000362 CM002911 EDX06007.1 KMY91971.1 CH940648 EDW60984.1 CH933808 EDW09988.1 CH954177 EDV59693.1 GEZM01097407 JAV54221.1 CH916367 EDW00943.1 KK852686 KDR18442.1 GAKP01011705 GAKP01011704 JAC47248.1 CH902619 EDV36435.1 OUUW01000001 SPP73140.1 CM000071 EAL24787.1 CH479181 EDW31498.1 CP012524 ALC40849.1 CCAG010005392 KB632197 ERL90018.1 JRES01001254 KNC24191.1 APGK01036978 KB740945 ENN77504.1 JXJN01011087 CH963849 EDW74609.1 UFQT01002915 SSX34272.1 KQ434864 KZC09085.1 GBYB01005731 JAG75498.1 CH477675 EAT37390.1 GGFK01010400 MBW43721.1 JXUM01066817 JXUM01066818 JXUM01066819 JXUM01066820 KQ562417 KXJ75955.1 KQ414613 KOC68986.1 KZ288269 PBC29963.1 GL441846 EFN64117.1 AXCM01001232 ATLV01022353 KE525331 KFB47558.1 ADMH02001049 ETN64313.1 GL450384 EFN81116.1 KQ978566 KYN30100.1 GFDL01014761 JAV20284.1 ADTU01004201 KQ976681 KYM78162.1 GFDL01014760 JAV20285.1 KQ977220 KYN04922.1 KQ981276 KYN43716.1 GL767043 EFZ14275.1 KQ982566 KYQ54729.1 GL888122 EGI66948.1 DS231921 EDS26526.1 LBMM01015162 KMQ84907.1 AXCN02002113 GGMS01013027 MBY82230.1 GFXV01000866 MBW12671.1 APCN01001944 GGMR01015232 MBY27851.1 GFDL01014828 JAV20217.1 GEDC01008738 JAS28560.1 KK107151 QOIP01000003 EZA57354.1 RLU24567.1 ABLF02039935 GL732685 EFX67138.1 LRGB01000512 KZS18370.1 GDIP01029553 JAM74162.1 GDIP01144285 JAJ79117.1 GDIQ01031482 JAN63255.1 DS235874 EEB19738.1 GDIQ01255518 JAJ96206.1 GDIP01045811 JAM57904.1 KK854471 PTY16132.1 AAAB01008966 EAA13035.4
PZC77561.1 NWSH01000828 PCG73967.1 KQ459193 KPJ03026.1 AGBW02014435 OWR41695.1 JTDY01006457 KOB65997.1 GAMC01002380 JAC04176.1 CM000157 EDW89176.1 KQ971354 EFA07079.1 AE013599 AY094923 AAF59259.1 AAM11276.1 CM000362 CM002911 EDX06007.1 KMY91971.1 CH940648 EDW60984.1 CH933808 EDW09988.1 CH954177 EDV59693.1 GEZM01097407 JAV54221.1 CH916367 EDW00943.1 KK852686 KDR18442.1 GAKP01011705 GAKP01011704 JAC47248.1 CH902619 EDV36435.1 OUUW01000001 SPP73140.1 CM000071 EAL24787.1 CH479181 EDW31498.1 CP012524 ALC40849.1 CCAG010005392 KB632197 ERL90018.1 JRES01001254 KNC24191.1 APGK01036978 KB740945 ENN77504.1 JXJN01011087 CH963849 EDW74609.1 UFQT01002915 SSX34272.1 KQ434864 KZC09085.1 GBYB01005731 JAG75498.1 CH477675 EAT37390.1 GGFK01010400 MBW43721.1 JXUM01066817 JXUM01066818 JXUM01066819 JXUM01066820 KQ562417 KXJ75955.1 KQ414613 KOC68986.1 KZ288269 PBC29963.1 GL441846 EFN64117.1 AXCM01001232 ATLV01022353 KE525331 KFB47558.1 ADMH02001049 ETN64313.1 GL450384 EFN81116.1 KQ978566 KYN30100.1 GFDL01014761 JAV20284.1 ADTU01004201 KQ976681 KYM78162.1 GFDL01014760 JAV20285.1 KQ977220 KYN04922.1 KQ981276 KYN43716.1 GL767043 EFZ14275.1 KQ982566 KYQ54729.1 GL888122 EGI66948.1 DS231921 EDS26526.1 LBMM01015162 KMQ84907.1 AXCN02002113 GGMS01013027 MBY82230.1 GFXV01000866 MBW12671.1 APCN01001944 GGMR01015232 MBY27851.1 GFDL01014828 JAV20217.1 GEDC01008738 JAS28560.1 KK107151 QOIP01000003 EZA57354.1 RLU24567.1 ABLF02039935 GL732685 EFX67138.1 LRGB01000512 KZS18370.1 GDIP01029553 JAM74162.1 GDIP01144285 JAJ79117.1 GDIQ01031482 JAN63255.1 DS235874 EEB19738.1 GDIQ01255518 JAJ96206.1 GDIP01045811 JAM57904.1 KK854471 PTY16132.1 AAAB01008966 EAA13035.4
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000037510
UP000095300
+ More
UP000002282 UP000007266 UP000000803 UP000092443 UP000000304 UP000008792 UP000009192 UP000008711 UP000095301 UP000192221 UP000001070 UP000027135 UP000007801 UP000092445 UP000091820 UP000268350 UP000001819 UP000008744 UP000092553 UP000092444 UP000030742 UP000037069 UP000192223 UP000019118 UP000092460 UP000007798 UP000076502 UP000008820 UP000002358 UP000069940 UP000249989 UP000053825 UP000242457 UP000000311 UP000075883 UP000030765 UP000000673 UP000069272 UP000005203 UP000008237 UP000078492 UP000075920 UP000005205 UP000078540 UP000078542 UP000078541 UP000075809 UP000007755 UP000002320 UP000036403 UP000075886 UP000075884 UP000075885 UP000075903 UP000075882 UP000075880 UP000075840 UP000075902 UP000076407 UP000075881 UP000053097 UP000279307 UP000007819 UP000000305 UP000075900 UP000076858 UP000009046 UP000076408 UP000075901 UP000007062
UP000002282 UP000007266 UP000000803 UP000092443 UP000000304 UP000008792 UP000009192 UP000008711 UP000095301 UP000192221 UP000001070 UP000027135 UP000007801 UP000092445 UP000091820 UP000268350 UP000001819 UP000008744 UP000092553 UP000092444 UP000030742 UP000037069 UP000192223 UP000019118 UP000092460 UP000007798 UP000076502 UP000008820 UP000002358 UP000069940 UP000249989 UP000053825 UP000242457 UP000000311 UP000075883 UP000030765 UP000000673 UP000069272 UP000005203 UP000008237 UP000078492 UP000075920 UP000005205 UP000078540 UP000078542 UP000078541 UP000075809 UP000007755 UP000002320 UP000036403 UP000075886 UP000075884 UP000075885 UP000075903 UP000075882 UP000075880 UP000075840 UP000075902 UP000076407 UP000075881 UP000053097 UP000279307 UP000007819 UP000000305 UP000075900 UP000076858 UP000009046 UP000076408 UP000075901 UP000007062
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9JHX4
A0A2W1BT90
A0A2A4JPI7
A0A194QHU4
A0A212EJN9
A0A0L7KRR8
+ More
W8BS29 A0A1I8PG43 B4P1P0 D6WS25 Q7JX95 A0A1A9XFG2 B4QE98 B4LMA6 B4KND5 B3N9Z7 A0A1Y1K169 A0A1I8M7W0 A0A1W4VD87 B4J6P0 A0A067R7I5 A0A034W0I0 B3MDQ0 A0A1B0AGG7 A0A1A9WR41 A0A3B0JD43 Q292S4 B4GCQ0 A0A0M3QUK8 A0A1B0G1H5 U4UCI4 A0A0L0BVZ0 A0A1W4WXZ4 N6TB60 A0A1B0BAP1 B4MR70 A0A336MV57 A0A154PB07 A0A0C9RFG5 Q16SD4 A0A2M4ASG8 K7IQP3 A0A182GIV0 A0A0L7RE32 A0A2A3EDV4 E2AQX1 A0A182MCU6 A0A084WBG4 W5JLY3 A0A182FTC1 A0A087ZYY8 E2BTB4 A0A151JSB8 A0A1Q3EYA6 A0A182WF41 A0A158NZA4 A0A195B1K1 A0A1Q3EYA3 A0A195CWL0 A0A195FTG4 E9IYS2 A0A151X374 F4WFQ5 B0WFZ1 A0A0J7K3J0 A0A182QVR7 A0A182MXY0 A0A2S2QXI9 A0A182P9E5 A0A2H8TF14 A0A182V1A3 A0A182LPK0 A0A182J894 A0A182HNM3 A0A182TTH8 A0A182XCC0 A0A2S2PEG3 A0A1Q3EY22 A0A1B6DSA6 A0A182K091 A0A026WQG8 J9JWR3 E9HMA4 A0A182RFC1 A0A162NY84 A0A0P6AGR6 A0A0P5EAK8 A0A0P6HAV8 E0W282 A0A0P5FYU1 A0A0P5ZDD5 A0A182YDC5 A0A182S8L5 A0A2R7W8U2 Q7Q318
W8BS29 A0A1I8PG43 B4P1P0 D6WS25 Q7JX95 A0A1A9XFG2 B4QE98 B4LMA6 B4KND5 B3N9Z7 A0A1Y1K169 A0A1I8M7W0 A0A1W4VD87 B4J6P0 A0A067R7I5 A0A034W0I0 B3MDQ0 A0A1B0AGG7 A0A1A9WR41 A0A3B0JD43 Q292S4 B4GCQ0 A0A0M3QUK8 A0A1B0G1H5 U4UCI4 A0A0L0BVZ0 A0A1W4WXZ4 N6TB60 A0A1B0BAP1 B4MR70 A0A336MV57 A0A154PB07 A0A0C9RFG5 Q16SD4 A0A2M4ASG8 K7IQP3 A0A182GIV0 A0A0L7RE32 A0A2A3EDV4 E2AQX1 A0A182MCU6 A0A084WBG4 W5JLY3 A0A182FTC1 A0A087ZYY8 E2BTB4 A0A151JSB8 A0A1Q3EYA6 A0A182WF41 A0A158NZA4 A0A195B1K1 A0A1Q3EYA3 A0A195CWL0 A0A195FTG4 E9IYS2 A0A151X374 F4WFQ5 B0WFZ1 A0A0J7K3J0 A0A182QVR7 A0A182MXY0 A0A2S2QXI9 A0A182P9E5 A0A2H8TF14 A0A182V1A3 A0A182LPK0 A0A182J894 A0A182HNM3 A0A182TTH8 A0A182XCC0 A0A2S2PEG3 A0A1Q3EY22 A0A1B6DSA6 A0A182K091 A0A026WQG8 J9JWR3 E9HMA4 A0A182RFC1 A0A162NY84 A0A0P6AGR6 A0A0P5EAK8 A0A0P6HAV8 E0W282 A0A0P5FYU1 A0A0P5ZDD5 A0A182YDC5 A0A182S8L5 A0A2R7W8U2 Q7Q318
PDB
5WZJ
E-value=8.81372e-22,
Score=258
Ontologies
GO
PANTHER
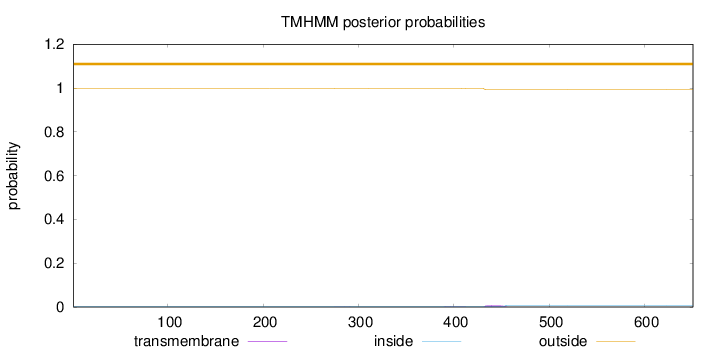
Topology
Length:
651
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.20462
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00217
outside
1 - 651
Population Genetic Test Statistics
Pi
21.209093
Theta
21.275732
Tajima's D
-1.601884
CLR
1.028156
CSRT
0.0455977201139943
Interpretation
Uncertain
