Gene
KWMTBOMO12296
Pre Gene Modal
BGIBMGA009158
Annotation
PREDICTED:_protein_CBFA2T1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.774
Sequence
CDS
ATGAATTATAAAACGGCAGCTCTGTCCTCAACGGAGTCCCTGTCTCCTCCTCTCGCTGATGGAGCACGGGCCCCGGCTCTGCAGCGGCTTCGGCGATTCCTCTCGGCTCTGCAACAGTTCGCCACGGACGTTGGTGCTGAAATCGGCGAGAGAGTTAAACATCTTATCCACAATCTTGTGGCAGGTACAATGAACGTCGAGGAGTTCCAGAGCGGCGTGCAGGAGTGCACCAACTACCCGCTGCGGGCCTCCGTGCCTGGCTTCCTCCGCGCGCTGCTGCCGCTAGCACAGCGAGACCTGCACGCGCGCGCCCGCAGAGCCAAGCAGACTCCCCTCCAGTACATCAGATCTCACGAGCACCTGATCTTGGAGTCAGGGGGAGACTCCAGCGACATATTCGCTCCGCAGCATCCCGGCACAGTTCCCGAAGGGGGCATCAAGAGACGAGCGAGTGATCCGTTCTACGAGACATCGCAAACAAACGGCGCACATCCCGAAGACTTCCCTCCTCACGCAAAAAGACCTCCCCCCTCCAGCTTATTCACGACCTCCCCCTTTTTATACCCTCTACCCAGCAACGCGAGTTTGTTCGAATACGGCCACTACCAAGGGTTTCATGCCGGACACAGTGATTCTGGATTTGAAAGAAGAGATGGTGGTATTTCGGTTCGAGATGTCTCCTCAATGAACGCGCCGTTCGAGCCGCGAGGTTCCAGTTCCTCGTCAGTCGGCTTACTCAAGACCGATGATGAGTGGAGGAACATCAACACCATGCTTAACTGTATCCTCAGCATGGTAGAGAAGACCAAACGGGCTTTGGCTATATTACAGCAAAGAGGAGTGGAGCCGCCAGAAAATAATGATATCAAAAGAACTGCAAACGAGATCATGGCGGCGGCCGTCCGGCAGACTGAAGAGCGAGTGGCAGAGGTTCGGAGACGGGCTGAAGATGCTGTCAATCAGGTAAAAAGGCAGGCTCTGATAGAGCTGCAGAGAGCCGTGGGTGCGGCGGAGAGCAAGGCGCTCGAGCTGGTGGCGCCGGCCGCCCCTCCCGCCGCGCCCGCCGCCCCGCCCGCACACCCCGCGCACCCCGCGCCGCCGCTCAGCCCCACTCCAGCGCCACACCATACGTGCTGCTGGAACTGCGGCCGGAAGGCTCAAGAGACGTGCTCTGGATGTAACTCGGCGCGGTACTGCGGCTCCTTCTGTCAGCACAAGGACTGGGAGAACCATCACCAGGTGTGCAGCGGACGTGACCAGAAGCCAACGGCTCTGATTCGAACCTCTCCCCCGACCACGCAACCCAGCCTGAAGCCGCGATCGACCACCCCGATAGCTGCCGCCGCCCAGCCCGACAGCCGCCTGTCCCTGAGCACGCTGGGCACGGCCGAGCGCCTGCTGACGGCGGAGCGCCCGCTGGCGACCGGCGCCGACCGCCTGGCGCTGGCGACTCTGTCAGCGGCCCAGAGTGCCGGCGGCGTCGGCACCAAGAAATGA
Protein
MNYKTAALSSTESLSPPLADGARAPALQRLRRFLSALQQFATDVGAEIGERVKHLIHNLVAGTMNVEEFQSGVQECTNYPLRASVPGFLRALLPLAQRDLHARARRAKQTPLQYIRSHEHLILESGGDSSDIFAPQHPGTVPEGGIKRRASDPFYETSQTNGAHPEDFPPHAKRPPPSSLFTTSPFLYPLPSNASLFEYGHYQGFHAGHSDSGFERRDGGISVRDVSSMNAPFEPRGSSSSSVGLLKTDDEWRNINTMLNCILSMVEKTKRALAILQQRGVEPPENNDIKRTANEIMAAAVRQTEERVAEVRRRAEDAVNQVKRQALIELQRAVGAAESKALELVAPAAPPAAPAAPPAHPAHPAPPLSPTPAPHHTCCWNCGRKAQETCSGCNSARYCGSFCQHKDWENHHQVCSGRDQKPTALIRTSPPTTQPSLKPRSTTPIAAAAQPDSRLSLSTLGTAERLLTAERPLATGADRLALATLSAAQSAGGVGTKK
Summary
Uniprot
A0A2H1W2X5
A0A2A4JPH4
A0A2W1BR36
A0A194QBU5
A0A0N1PFR6
A0A212EJM9
+ More
A0A139WEF7 A0A1W4WLX1 U5EYB2 A0A336MKV4 A0A1Y1KHN6 A0A182W2C0 A0A2M4BIW2 Q16H02 A0A2M4BI68 A0A2M3ZKS2 W5JJJ4 A0A182QQD8 A0A182GIY3 A0A182R577 A0A2M4AKI2 A0A2M4AK15 A0A2M4AI29 A0A182FVQ8 A0A2M4AHU2 A0A182GFI4 A0A182YMH0 A0A084WUR2 A0A182JYU7 A0A087T4P0 A0A1J1HI95 A0A1L8DDI8 A0A3Q2TJV8 C3ZBL4 A0A3B3WXJ5 A0A3B4F6X1 L5JNJ9 H2U0M9 A0A1I8MYU8 A0A3Q1JWW0 A0A3P9J989 A0A3P9HT63 A0A3P9L2M1 A0A3P9MDM5 H2LNE5 A0A3Q1I964 A0A3B4AYS7 A0A3Q1BR38 A0A1A9VQ65 A0A1B0ADB6 A0A3B3CEB1 A0A3B3CED4 H2U0N1 A0A3S2PY62 A0A3B3CC25 A0A2R8PVW8 A0A1A9XF01 H2U0M8 A0A1I8PZF4 A0A1I8PZH7 A0A3Q0SUZ8 A0A1I8PZJ9 A0A2K6RKK1 Q5IJ71 A0A0L0BPF6 A0A2I4ASR7 A0A3B4CJC9 A0A1A9W8E6 A0A3P8TWZ4 A0A3B4TUE1 A0A3Q4HX33 A0A3P8U2Q7 N6U771 A0A3Q2YN62 A0A3Q2XEY3 A0A1I8PZH6 A0A3Q1BN58 A0A3Q7WIV7 A0A3Q1GRI4 A0A2R8Q304 A0A3B3CEF9 A0A3B3CCQ1 A0A096LZH3 A0A2U9BEU2 A0A2R8Q678 A0A3B5APC7 A0A3Q3JA68 A0A1B0D6E2 A0A3Q2PNV7 A0A3B3CG71 A0A1A8FDK5 A0A3B5A2R3 A0A1A8NBL0 A0A1A8AYE3 A0A1A8S6Z0 A0A1A8J1X6 A0A1A8BIK2 A0A1A8S6R9
A0A139WEF7 A0A1W4WLX1 U5EYB2 A0A336MKV4 A0A1Y1KHN6 A0A182W2C0 A0A2M4BIW2 Q16H02 A0A2M4BI68 A0A2M3ZKS2 W5JJJ4 A0A182QQD8 A0A182GIY3 A0A182R577 A0A2M4AKI2 A0A2M4AK15 A0A2M4AI29 A0A182FVQ8 A0A2M4AHU2 A0A182GFI4 A0A182YMH0 A0A084WUR2 A0A182JYU7 A0A087T4P0 A0A1J1HI95 A0A1L8DDI8 A0A3Q2TJV8 C3ZBL4 A0A3B3WXJ5 A0A3B4F6X1 L5JNJ9 H2U0M9 A0A1I8MYU8 A0A3Q1JWW0 A0A3P9J989 A0A3P9HT63 A0A3P9L2M1 A0A3P9MDM5 H2LNE5 A0A3Q1I964 A0A3B4AYS7 A0A3Q1BR38 A0A1A9VQ65 A0A1B0ADB6 A0A3B3CEB1 A0A3B3CED4 H2U0N1 A0A3S2PY62 A0A3B3CC25 A0A2R8PVW8 A0A1A9XF01 H2U0M8 A0A1I8PZF4 A0A1I8PZH7 A0A3Q0SUZ8 A0A1I8PZJ9 A0A2K6RKK1 Q5IJ71 A0A0L0BPF6 A0A2I4ASR7 A0A3B4CJC9 A0A1A9W8E6 A0A3P8TWZ4 A0A3B4TUE1 A0A3Q4HX33 A0A3P8U2Q7 N6U771 A0A3Q2YN62 A0A3Q2XEY3 A0A1I8PZH6 A0A3Q1BN58 A0A3Q7WIV7 A0A3Q1GRI4 A0A2R8Q304 A0A3B3CEF9 A0A3B3CCQ1 A0A096LZH3 A0A2U9BEU2 A0A2R8Q678 A0A3B5APC7 A0A3Q3JA68 A0A1B0D6E2 A0A3Q2PNV7 A0A3B3CG71 A0A1A8FDK5 A0A3B5A2R3 A0A1A8NBL0 A0A1A8AYE3 A0A1A8S6Z0 A0A1A8J1X6 A0A1A8BIK2 A0A1A8S6R9
Pubmed
EMBL
ODYU01005959
SOQ47387.1
NWSH01000828
PCG73965.1
KZ149926
PZC77559.1
+ More
KQ459193 KPJ03028.1 KQ461072 KPJ09829.1 AGBW02014435 OWR41697.1 KQ971354 KYB26340.1 GANO01002108 JAB57763.1 UFQT01001350 SSX30161.1 GEZM01088251 JAV58277.1 GGFJ01003846 MBW52987.1 CH478219 EAT33521.1 GGFJ01003367 MBW52508.1 GGFM01008383 MBW29134.1 ADMH02001301 ETN63075.1 AXCN02001124 JXUM01067025 JXUM01067026 JXUM01067027 JXUM01067028 JXUM01067029 JXUM01067030 KQ562429 KXJ75928.1 GGFK01007972 MBW41293.1 GGFK01007809 MBW41130.1 GGFK01007086 MBW40407.1 GGFK01007020 MBW40341.1 JXUM01010513 JXUM01010514 JXUM01010515 KQ560310 KXJ83118.1 ATLV01027104 ATLV01027105 KE525423 KFB53956.1 KK113388 KFM60079.1 CVRI01000004 CRK87132.1 GFDF01009684 JAV04400.1 GG666603 EEN50251.1 KB031158 ELK00507.1 CM012441 RVE73081.1 AL807378 BX248306 AY714078 AAW49216.1 JRES01001654 KNC21114.1 APGK01036968 APGK01036969 APGK01036970 KB740945 ENN77500.1 AYCK01000329 AYCK01000330 CP026248 AWP02515.1 AJVK01025937 AJVK01025938 HAEB01010948 HAEC01006838 SBQ57475.1 HAEG01001926 SBR66232.1 HADY01021549 SBP60034.1 HAEI01011743 SBS14115.1 HAED01017220 HAEE01009437 SBR03665.1 HADZ01002389 HAEA01010189 SBP66330.1 HAEH01022431 SBS13951.1
KQ459193 KPJ03028.1 KQ461072 KPJ09829.1 AGBW02014435 OWR41697.1 KQ971354 KYB26340.1 GANO01002108 JAB57763.1 UFQT01001350 SSX30161.1 GEZM01088251 JAV58277.1 GGFJ01003846 MBW52987.1 CH478219 EAT33521.1 GGFJ01003367 MBW52508.1 GGFM01008383 MBW29134.1 ADMH02001301 ETN63075.1 AXCN02001124 JXUM01067025 JXUM01067026 JXUM01067027 JXUM01067028 JXUM01067029 JXUM01067030 KQ562429 KXJ75928.1 GGFK01007972 MBW41293.1 GGFK01007809 MBW41130.1 GGFK01007086 MBW40407.1 GGFK01007020 MBW40341.1 JXUM01010513 JXUM01010514 JXUM01010515 KQ560310 KXJ83118.1 ATLV01027104 ATLV01027105 KE525423 KFB53956.1 KK113388 KFM60079.1 CVRI01000004 CRK87132.1 GFDF01009684 JAV04400.1 GG666603 EEN50251.1 KB031158 ELK00507.1 CM012441 RVE73081.1 AL807378 BX248306 AY714078 AAW49216.1 JRES01001654 KNC21114.1 APGK01036968 APGK01036969 APGK01036970 KB740945 ENN77500.1 AYCK01000329 AYCK01000330 CP026248 AWP02515.1 AJVK01025937 AJVK01025938 HAEB01010948 HAEC01006838 SBQ57475.1 HAEG01001926 SBR66232.1 HADY01021549 SBP60034.1 HAEI01011743 SBS14115.1 HAED01017220 HAEE01009437 SBR03665.1 HADZ01002389 HAEA01010189 SBP66330.1 HAEH01022431 SBS13951.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000007266
UP000192223
+ More
UP000075920 UP000008820 UP000000673 UP000075886 UP000069940 UP000249989 UP000075900 UP000069272 UP000076408 UP000030765 UP000075881 UP000054359 UP000183832 UP000265000 UP000001554 UP000261480 UP000261460 UP000010552 UP000005226 UP000095301 UP000265040 UP000265200 UP000265180 UP000001038 UP000261520 UP000257160 UP000078200 UP000092445 UP000261560 UP000000437 UP000092443 UP000095300 UP000261340 UP000233200 UP000037069 UP000192220 UP000261440 UP000091820 UP000265080 UP000261420 UP000261580 UP000019118 UP000264820 UP000286642 UP000257200 UP000028760 UP000246464 UP000261400 UP000261600 UP000092462
UP000075920 UP000008820 UP000000673 UP000075886 UP000069940 UP000249989 UP000075900 UP000069272 UP000076408 UP000030765 UP000075881 UP000054359 UP000183832 UP000265000 UP000001554 UP000261480 UP000261460 UP000010552 UP000005226 UP000095301 UP000265040 UP000265200 UP000265180 UP000001038 UP000261520 UP000257160 UP000078200 UP000092445 UP000261560 UP000000437 UP000092443 UP000095300 UP000261340 UP000233200 UP000037069 UP000192220 UP000261440 UP000091820 UP000265080 UP000261420 UP000261580 UP000019118 UP000264820 UP000286642 UP000257200 UP000028760 UP000246464 UP000261400 UP000261600 UP000092462
Interpro
Gene 3D
ProteinModelPortal
A0A2H1W2X5
A0A2A4JPH4
A0A2W1BR36
A0A194QBU5
A0A0N1PFR6
A0A212EJM9
+ More
A0A139WEF7 A0A1W4WLX1 U5EYB2 A0A336MKV4 A0A1Y1KHN6 A0A182W2C0 A0A2M4BIW2 Q16H02 A0A2M4BI68 A0A2M3ZKS2 W5JJJ4 A0A182QQD8 A0A182GIY3 A0A182R577 A0A2M4AKI2 A0A2M4AK15 A0A2M4AI29 A0A182FVQ8 A0A2M4AHU2 A0A182GFI4 A0A182YMH0 A0A084WUR2 A0A182JYU7 A0A087T4P0 A0A1J1HI95 A0A1L8DDI8 A0A3Q2TJV8 C3ZBL4 A0A3B3WXJ5 A0A3B4F6X1 L5JNJ9 H2U0M9 A0A1I8MYU8 A0A3Q1JWW0 A0A3P9J989 A0A3P9HT63 A0A3P9L2M1 A0A3P9MDM5 H2LNE5 A0A3Q1I964 A0A3B4AYS7 A0A3Q1BR38 A0A1A9VQ65 A0A1B0ADB6 A0A3B3CEB1 A0A3B3CED4 H2U0N1 A0A3S2PY62 A0A3B3CC25 A0A2R8PVW8 A0A1A9XF01 H2U0M8 A0A1I8PZF4 A0A1I8PZH7 A0A3Q0SUZ8 A0A1I8PZJ9 A0A2K6RKK1 Q5IJ71 A0A0L0BPF6 A0A2I4ASR7 A0A3B4CJC9 A0A1A9W8E6 A0A3P8TWZ4 A0A3B4TUE1 A0A3Q4HX33 A0A3P8U2Q7 N6U771 A0A3Q2YN62 A0A3Q2XEY3 A0A1I8PZH6 A0A3Q1BN58 A0A3Q7WIV7 A0A3Q1GRI4 A0A2R8Q304 A0A3B3CEF9 A0A3B3CCQ1 A0A096LZH3 A0A2U9BEU2 A0A2R8Q678 A0A3B5APC7 A0A3Q3JA68 A0A1B0D6E2 A0A3Q2PNV7 A0A3B3CG71 A0A1A8FDK5 A0A3B5A2R3 A0A1A8NBL0 A0A1A8AYE3 A0A1A8S6Z0 A0A1A8J1X6 A0A1A8BIK2 A0A1A8S6R9
A0A139WEF7 A0A1W4WLX1 U5EYB2 A0A336MKV4 A0A1Y1KHN6 A0A182W2C0 A0A2M4BIW2 Q16H02 A0A2M4BI68 A0A2M3ZKS2 W5JJJ4 A0A182QQD8 A0A182GIY3 A0A182R577 A0A2M4AKI2 A0A2M4AK15 A0A2M4AI29 A0A182FVQ8 A0A2M4AHU2 A0A182GFI4 A0A182YMH0 A0A084WUR2 A0A182JYU7 A0A087T4P0 A0A1J1HI95 A0A1L8DDI8 A0A3Q2TJV8 C3ZBL4 A0A3B3WXJ5 A0A3B4F6X1 L5JNJ9 H2U0M9 A0A1I8MYU8 A0A3Q1JWW0 A0A3P9J989 A0A3P9HT63 A0A3P9L2M1 A0A3P9MDM5 H2LNE5 A0A3Q1I964 A0A3B4AYS7 A0A3Q1BR38 A0A1A9VQ65 A0A1B0ADB6 A0A3B3CEB1 A0A3B3CED4 H2U0N1 A0A3S2PY62 A0A3B3CC25 A0A2R8PVW8 A0A1A9XF01 H2U0M8 A0A1I8PZF4 A0A1I8PZH7 A0A3Q0SUZ8 A0A1I8PZJ9 A0A2K6RKK1 Q5IJ71 A0A0L0BPF6 A0A2I4ASR7 A0A3B4CJC9 A0A1A9W8E6 A0A3P8TWZ4 A0A3B4TUE1 A0A3Q4HX33 A0A3P8U2Q7 N6U771 A0A3Q2YN62 A0A3Q2XEY3 A0A1I8PZH6 A0A3Q1BN58 A0A3Q7WIV7 A0A3Q1GRI4 A0A2R8Q304 A0A3B3CEF9 A0A3B3CCQ1 A0A096LZH3 A0A2U9BEU2 A0A2R8Q678 A0A3B5APC7 A0A3Q3JA68 A0A1B0D6E2 A0A3Q2PNV7 A0A3B3CG71 A0A1A8FDK5 A0A3B5A2R3 A0A1A8NBL0 A0A1A8AYE3 A0A1A8S6Z0 A0A1A8J1X6 A0A1A8BIK2 A0A1A8S6R9
PDB
5ECJ
E-value=4.13277e-23,
Score=268
Ontologies
GO
PANTHER
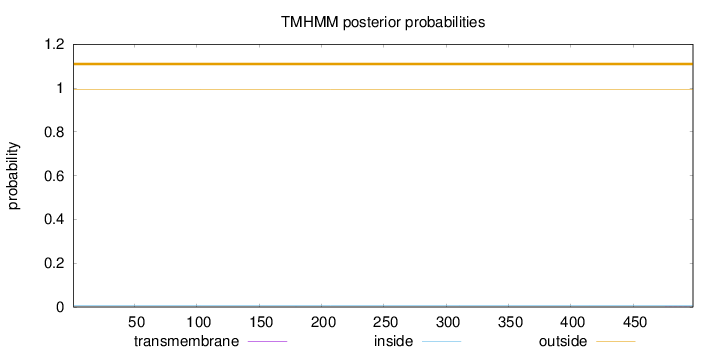
Topology
Length:
498
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00565
Exp number, first 60 AAs:
0.000580000000000001
Total prob of N-in:
0.00543
outside
1 - 498
Population Genetic Test Statistics
Pi
22.362994
Theta
18.082947
Tajima's D
-1.277413
CLR
0.746689
CSRT
0.0889955502224889
Interpretation
Uncertain
