Gene
KWMTBOMO12290
Pre Gene Modal
BGIBMGA009126
Annotation
PREDICTED:_oxidative_stress-induced_growth_inhibitor_1-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.78
Sequence
CDS
ATGAAGGACAGCATGAGGAGTTGTGAACACGTGCTGACAGACGATGTGGAGTACAAGGAAGTGGTCGTGATAGGAAATGGGCCCAGCGGTATGGTGACCTCATTCATGCTGGACGGCAATGTACCATACCTCAAACAGATACCAGAAGACTTGCCAATTGACGAGATGCTCAGAGCCAGACTTTCAAACCTTCCCGAAGGCCAGAGCCTGTACGACGCAGACCTCGCGGAGTTGGCCGAGGGATTGGAAGGCAGAAGTCAAAACCCGATACCTATATTGATGGACGCTCTCCTGAGGCCTTGCGCGGACCTGGGCGTCCAGGCCGACTCGCTCATCGAGTGGCGCTACGAGAGCGACAAGCAGATAGACCATGTGGTGTTAGGGCAGGGCCCGCCGGGGGGCGCCTGGCACACGTTCCCGCCCGACGTGCGCACGCTGTCCCCGGCGGCCTGGCTCACGCTGCCCCCGCACACGGACGGCGGCGCGGAGCGGCTGTCGGCGCGGGCCGTCGCCAACTACTGCAGGAGATACGTGCACGCCTGCAAGCTGCAGGTATGGACCCCCGCCACCGTTGCACCCTGGATGGGCTCTCTAGCCACCACTGAACCCTGGATAGGTTCTCTAGTAAACGTTTCGTGA
Protein
MKDSMRSCEHVLTDDVEYKEVVVIGNGPSGMVTSFMLDGNVPYLKQIPEDLPIDEMLRARLSNLPEGQSLYDADLAELAEGLEGRSQNPIPILMDALLRPCADLGVQADSLIEWRYESDKQIDHVVLGQGPPGGAWHTFPPDVRTLSPAAWLTLPPHTDGGAERLSARAVANYCRRYVHACKLQVWTPATVAPWMGSLATTEPWIGSLVNVS
Summary
Uniprot
A0A3S2P6A6
A0A0N1PGZ3
A0A194QC39
A0A212FJZ2
A0A182LPS3
A0A182HNR5
+ More
A0A182MFR3 A0A182J2V3 A0A182WG15 A0A1Y1N9G7 A0A1Y1N9G6 A0A182FVR7 A0A182MY86 A0A182H8F6 A0A023EV31 A0A182PTT7 A0A182U433 A0A1B6D3R4 A0A1B0CRY7 A0A182V7T0 A0A1L8DD72 A0A2P8Y868 A0A1B6FJL1 A0A1B6I5I8 R4FR86 A0A0P4VRN2 A0A224XSK8 A0A023F8F3 A0A0V0GAR7 A0A1W4X2X8 A0A0T6BG65 A0A2J7PNN9 A0A226EPY7 A0A1Z5LBL1 A0A1D2N165 A0A1A7WTX6 A0A1A8RPE0 A0A1A8R442 A0A1A8IZI6 A0A1A8C1V5 G1MQ40 A0A1A8L7W6 A0A1A8G9G9 L8I4H0 A0A3B3C5N3 A0A1A8KQ42 A0A0P7V006 W5PJ78 A0A146XX09 A0A1U8D671 A0A3Q1HG82 A0A3B4DT93 A0A096LT90 Q2KIN7 A0A1S3EY60 M3XDA0 I3J9H7 A0A151MLN5 A0A1Z5L6S6 A0A2I4CIR5 A0A3Q4MUD6 A0A3B3WNS2 A0A087Y1X8 A0A3B3W3E7 A0A3P8P8I4 A0A3P9CI33 A0A3B4G1L5 A0A154PAY3 A0A1A8ATL4 T1J4X7 A0A3P9QHH2 A0A3Q2GP17
A0A182MFR3 A0A182J2V3 A0A182WG15 A0A1Y1N9G7 A0A1Y1N9G6 A0A182FVR7 A0A182MY86 A0A182H8F6 A0A023EV31 A0A182PTT7 A0A182U433 A0A1B6D3R4 A0A1B0CRY7 A0A182V7T0 A0A1L8DD72 A0A2P8Y868 A0A1B6FJL1 A0A1B6I5I8 R4FR86 A0A0P4VRN2 A0A224XSK8 A0A023F8F3 A0A0V0GAR7 A0A1W4X2X8 A0A0T6BG65 A0A2J7PNN9 A0A226EPY7 A0A1Z5LBL1 A0A1D2N165 A0A1A7WTX6 A0A1A8RPE0 A0A1A8R442 A0A1A8IZI6 A0A1A8C1V5 G1MQ40 A0A1A8L7W6 A0A1A8G9G9 L8I4H0 A0A3B3C5N3 A0A1A8KQ42 A0A0P7V006 W5PJ78 A0A146XX09 A0A1U8D671 A0A3Q1HG82 A0A3B4DT93 A0A096LT90 Q2KIN7 A0A1S3EY60 M3XDA0 I3J9H7 A0A151MLN5 A0A1Z5L6S6 A0A2I4CIR5 A0A3Q4MUD6 A0A3B3WNS2 A0A087Y1X8 A0A3B3W3E7 A0A3P8P8I4 A0A3P9CI33 A0A3B4G1L5 A0A154PAY3 A0A1A8ATL4 T1J4X7 A0A3P9QHH2 A0A3Q2GP17
Pubmed
EMBL
RSAL01000012
RVE53456.1
KQ461072
KPJ09824.1
KQ459193
KPJ03032.1
+ More
AGBW02008171 OWR54063.1 APCN01001971 AXCM01005958 GEZM01009493 GEZM01009492 JAV94562.1 GEZM01009494 JAV94561.1 JXUM01029045 JXUM01029046 KQ560840 KXJ80659.1 GAPW01000288 JAC13310.1 GEDC01016959 JAS20339.1 AJWK01025482 AJWK01025483 GFDF01009820 JAV04264.1 PYGN01000811 PSN40456.1 GECZ01019566 GECZ01004285 JAS50203.1 JAS65484.1 GECU01025539 GECU01009704 JAS82167.1 JAS98002.1 GAHY01000044 JAA77466.1 GDKW01002055 JAI54540.1 GFTR01005465 JAW10961.1 GBBI01001117 JAC17595.1 GECL01001167 JAP04957.1 LJIG01000599 KRT86341.1 NEVH01023952 PNF17931.1 LNIX01000002 OXA58881.1 GFJQ02002300 JAW04670.1 LJIJ01000306 ODM98978.1 HADW01007793 SBP09193.1 HAEH01018062 SBS07109.1 HAEI01007190 SBS00746.1 HAED01016334 SBR02779.1 HADZ01008895 HAEA01009726 SBP72836.1 HAEF01002592 SBR39974.1 HAEB01021154 HAEC01005459 SBQ67681.1 JH882006 ELR51405.1 HAEE01014271 SBR34321.1 JARO02005005 KPP67507.1 AMGL01027220 AMGL01027221 GCES01040428 JAR45895.1 AYCK01017041 AYCK01017042 BC112571 AAI12572.1 AANG04002512 AERX01019607 AKHW03005795 KYO25435.1 GFJQ02004072 JAW02898.1 KQ434864 KZC09069.1 HADY01019997 HAEJ01014376 SBP58482.1 JH431850
AGBW02008171 OWR54063.1 APCN01001971 AXCM01005958 GEZM01009493 GEZM01009492 JAV94562.1 GEZM01009494 JAV94561.1 JXUM01029045 JXUM01029046 KQ560840 KXJ80659.1 GAPW01000288 JAC13310.1 GEDC01016959 JAS20339.1 AJWK01025482 AJWK01025483 GFDF01009820 JAV04264.1 PYGN01000811 PSN40456.1 GECZ01019566 GECZ01004285 JAS50203.1 JAS65484.1 GECU01025539 GECU01009704 JAS82167.1 JAS98002.1 GAHY01000044 JAA77466.1 GDKW01002055 JAI54540.1 GFTR01005465 JAW10961.1 GBBI01001117 JAC17595.1 GECL01001167 JAP04957.1 LJIG01000599 KRT86341.1 NEVH01023952 PNF17931.1 LNIX01000002 OXA58881.1 GFJQ02002300 JAW04670.1 LJIJ01000306 ODM98978.1 HADW01007793 SBP09193.1 HAEH01018062 SBS07109.1 HAEI01007190 SBS00746.1 HAED01016334 SBR02779.1 HADZ01008895 HAEA01009726 SBP72836.1 HAEF01002592 SBR39974.1 HAEB01021154 HAEC01005459 SBQ67681.1 JH882006 ELR51405.1 HAEE01014271 SBR34321.1 JARO02005005 KPP67507.1 AMGL01027220 AMGL01027221 GCES01040428 JAR45895.1 AYCK01017041 AYCK01017042 BC112571 AAI12572.1 AANG04002512 AERX01019607 AKHW03005795 KYO25435.1 GFJQ02004072 JAW02898.1 KQ434864 KZC09069.1 HADY01019997 HAEJ01014376 SBP58482.1 JH431850
Proteomes
UP000283053
UP000053240
UP000053268
UP000007151
UP000075882
UP000075840
+ More
UP000075883 UP000075880 UP000075920 UP000069272 UP000075884 UP000069940 UP000249989 UP000075885 UP000075902 UP000092461 UP000075903 UP000245037 UP000192223 UP000235965 UP000198287 UP000094527 UP000001645 UP000261560 UP000034805 UP000002356 UP000189705 UP000265040 UP000261440 UP000028760 UP000081671 UP000011712 UP000005207 UP000050525 UP000192220 UP000261580 UP000261480 UP000261500 UP000265100 UP000265160 UP000261460 UP000076502 UP000242638 UP000265020
UP000075883 UP000075880 UP000075920 UP000069272 UP000075884 UP000069940 UP000249989 UP000075885 UP000075902 UP000092461 UP000075903 UP000245037 UP000192223 UP000235965 UP000198287 UP000094527 UP000001645 UP000261560 UP000034805 UP000002356 UP000189705 UP000265040 UP000261440 UP000028760 UP000081671 UP000011712 UP000005207 UP000050525 UP000192220 UP000261580 UP000261480 UP000261500 UP000265100 UP000265160 UP000261460 UP000076502 UP000242638 UP000265020
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
A0A3S2P6A6
A0A0N1PGZ3
A0A194QC39
A0A212FJZ2
A0A182LPS3
A0A182HNR5
+ More
A0A182MFR3 A0A182J2V3 A0A182WG15 A0A1Y1N9G7 A0A1Y1N9G6 A0A182FVR7 A0A182MY86 A0A182H8F6 A0A023EV31 A0A182PTT7 A0A182U433 A0A1B6D3R4 A0A1B0CRY7 A0A182V7T0 A0A1L8DD72 A0A2P8Y868 A0A1B6FJL1 A0A1B6I5I8 R4FR86 A0A0P4VRN2 A0A224XSK8 A0A023F8F3 A0A0V0GAR7 A0A1W4X2X8 A0A0T6BG65 A0A2J7PNN9 A0A226EPY7 A0A1Z5LBL1 A0A1D2N165 A0A1A7WTX6 A0A1A8RPE0 A0A1A8R442 A0A1A8IZI6 A0A1A8C1V5 G1MQ40 A0A1A8L7W6 A0A1A8G9G9 L8I4H0 A0A3B3C5N3 A0A1A8KQ42 A0A0P7V006 W5PJ78 A0A146XX09 A0A1U8D671 A0A3Q1HG82 A0A3B4DT93 A0A096LT90 Q2KIN7 A0A1S3EY60 M3XDA0 I3J9H7 A0A151MLN5 A0A1Z5L6S6 A0A2I4CIR5 A0A3Q4MUD6 A0A3B3WNS2 A0A087Y1X8 A0A3B3W3E7 A0A3P8P8I4 A0A3P9CI33 A0A3B4G1L5 A0A154PAY3 A0A1A8ATL4 T1J4X7 A0A3P9QHH2 A0A3Q2GP17
A0A182MFR3 A0A182J2V3 A0A182WG15 A0A1Y1N9G7 A0A1Y1N9G6 A0A182FVR7 A0A182MY86 A0A182H8F6 A0A023EV31 A0A182PTT7 A0A182U433 A0A1B6D3R4 A0A1B0CRY7 A0A182V7T0 A0A1L8DD72 A0A2P8Y868 A0A1B6FJL1 A0A1B6I5I8 R4FR86 A0A0P4VRN2 A0A224XSK8 A0A023F8F3 A0A0V0GAR7 A0A1W4X2X8 A0A0T6BG65 A0A2J7PNN9 A0A226EPY7 A0A1Z5LBL1 A0A1D2N165 A0A1A7WTX6 A0A1A8RPE0 A0A1A8R442 A0A1A8IZI6 A0A1A8C1V5 G1MQ40 A0A1A8L7W6 A0A1A8G9G9 L8I4H0 A0A3B3C5N3 A0A1A8KQ42 A0A0P7V006 W5PJ78 A0A146XX09 A0A1U8D671 A0A3Q1HG82 A0A3B4DT93 A0A096LT90 Q2KIN7 A0A1S3EY60 M3XDA0 I3J9H7 A0A151MLN5 A0A1Z5L6S6 A0A2I4CIR5 A0A3Q4MUD6 A0A3B3WNS2 A0A087Y1X8 A0A3B3W3E7 A0A3P8P8I4 A0A3P9CI33 A0A3B4G1L5 A0A154PAY3 A0A1A8ATL4 T1J4X7 A0A3P9QHH2 A0A3Q2GP17
Ontologies
GO
PANTHER
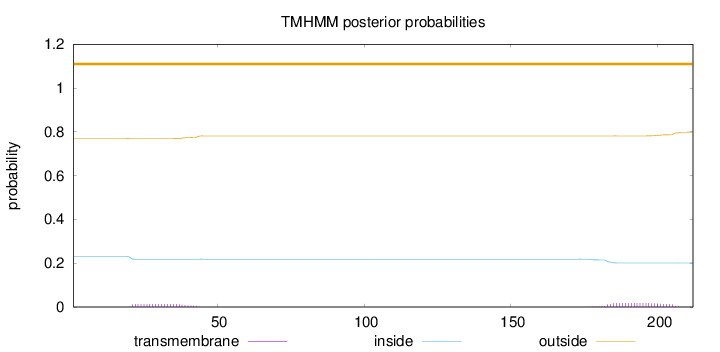
Topology
Length:
212
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.66984
Exp number, first 60 AAs:
0.26726
Total prob of N-in:
0.23080
outside
1 - 212
Population Genetic Test Statistics
Pi
29.121588
Theta
26.275906
Tajima's D
0.190125
CLR
0.518492
CSRT
0.421928903554822
Interpretation
Uncertain
