Gene
KWMTBOMO12286 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009132
Annotation
beta-tubulin_[Bombyx_mori]
Full name
Tubulin beta chain
+ More
Tubulin beta-1 chain
Tubulin beta-1 chain
Alternative Name
Beta-1-tubulin
Location in the cell
Cytoplasmic Reliability : 3.706
Sequence
CDS
ATGAGGGAAATCGTTCATCTACAGGCCGGCCAATGTGGTAACCAGATTGGAGCTAAGTTCTGGGAGATCATCTCCGACGAGCACGGCATCGACCCCACCGGTGCCTACCATGGAGACTCTGACTTGCAGTTGGAGCGCATCAATGTATACTACAATGAAGCCTCCGGCGGCAAGTACGTGCCCCGCGCCATCCTCGTCGACTTGGAGCCCGGCACCATGGACTCTGTCCGCTCCGGACCTTTCGGACAGATCTTCCGTCCGGACAACTTCGTCTTCGGACAGTCCGGCGCCGGCAACAACTGGGCCAAGGGACACTACACAGAGGGTGCTGAGCTCGTTGACTCGGTCCTCGATGTAGTCCGCAAAGAATCAGAATCTTGCGATTGCCTACAGGGCTTCCAACTTACACATTCCCTCGGTGGCGGCACCGGGTCCGGTATGGGCACCCTCCTCATCTCAAAGATCCGTGAAGAGTACCCCGACAGAATCATGAACACATACTCAGTAGTCCCCTCGCCCAAAGTATCAGACACTGTCGTCGAACCATACAATGCGACTCTCTCAGTTCACCAGCTAGTTGAAAACACAGACGAAACATACTGCATCGACAACGAGGCTCTATACGACATCTGCTTCCGCACTCTCAAACTGTCCACACCCACTTACGGTGACCTTAACCACTTAGTTTCCCTCACAATGTCTGGTGTCACCACTTGCCTTAGGTTCCCCGGTCAGTTGAACGCTGATCTCAGAAAATTGGCGGTAAACATGGTTCCCTTCCCGCGTCTCCACTTCTTCATGCCAGGTTTCGCTCCCCTCACATCCCGTGGAAGCAGACAGTACCGTGCCTTGACTGTACCCGAGCTCACGCAACAGATGTTCGACGCCAAGAACATGATGGCTGCCTGCGACCCACGCCACGGCCGCTACCTCACCGTCGCCGCCATCTTCCGCGGCCGCATGTCCATGAAGGAGGTCGACGAACAAATGCTTAACATCCAGAACAAGAACTCCTCCTACTTCGTGGAATGGATCCCGAACAACGTGAAGACCGCCGTGTGCGACATTCCTCCTCGTGGTCTCAAGATGGCCGCCACCTTCATCGGAAACTCCACCGCCATCCAGGAGCTGTTCAAGCGCATCTCGGAACAGTTCACCGCTATGTTCAGGCGCAAGGCTTTCTTGCATTGGTACACCGGCGAGGGCATGGACGAGATGGAGTTCACCGAGGCTGAGAGCAACATGAACGACCTTGTCTCCGAGTACCAACAGTACCAGGAGGCCACCGCCGACGAGGACGCAGAGTTTGACGAAGAGCAGGAGCAGGAGATTGAAGAGCATTAA
Protein
MREIVHLQAGQCGNQIGAKFWEIISDEHGIDPTGAYHGDSDLQLERINVYYNEASGGKYVPRAILVDLEPGTMDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVVRKESESCDCLQGFQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTYSVVPSPKVSDTVVEPYNATLSVHQLVENTDETYCIDNEALYDICFRTLKLSTPTYGDLNHLVSLTMSGVTTCLRFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSRQYRALTVPELTQQMFDAKNMMAACDPRHGRYLTVAAIFRGRMSMKEVDEQMLNIQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMAATFIGNSTAIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQEATADEDAEFDEEQEQEIEEH
Summary
Description
Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain (By similarity).
Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain (By similarity).
Subunit
Dimer of alpha and beta chains. A typical microtubule is a hollow water-filled tube with an outer diameter of 25 nm and an inner diameter of 15 nM. Alpha-beta heterodimers associate head-to-tail to form protofilaments running lengthwise along the microtubule wall with the beta-tubulin subunit facing the microtubule plus end conferring a structural polarity. Microtubules usually have 13 protofilaments but different protofilament numbers can be found in some organisms and specialized cells.
Dimer of alpha and beta chains. A typical microtubule is a hollow water-filled tube with an outer diameter of 25 nm and an inner diameter of 15 nM. Alpha-beta heterodimers associate head-to-tail to form protofilaments running lengthwise along the microtubule wall with the beta-tubulin subunit facing the microtubule plus end conferring a structural polarity. Microtubules usually have 13 protofilaments but different protofilament numbers can be found in some organisms and specialized cells. Interacts with mgr and Vhl.
Dimer of alpha and beta chains. A typical microtubule is a hollow water-filled tube with an outer diameter of 25 nm and an inner diameter of 15 nM. Alpha-beta heterodimers associate head-to-tail to form protofilaments running lengthwise along the microtubule wall with the beta-tubulin subunit facing the microtubule plus end conferring a structural polarity. Microtubules usually have 13 protofilaments but different protofilament numbers can be found in some organisms and specialized cells. Interacts with mgr and Vhl.
Similarity
Belongs to the tubulin family.
Keywords
Cytoplasm
Cytoskeleton
GTP-binding
Microtubule
Nucleotide-binding
Complete proteome
Phosphoprotein
Reference proteome
Feature
chain Tubulin beta-1 chain
Uniprot
A0A3S2NZY9
A0A2W1BMR7
I4DM38
Q8T8B3
A0A194QHV4
A0A1E1WVR1
+ More
O01676 A0A2Z5T6R2 I4DMM6 A0A3S2M7U1 A0A1E1W3A9 I4DIS9 Q8T8B2 A9YLN3 A0A212FJY5 F6M9L9 G1E6C4 A0A2J7QIE5 A0A1Y1KNW8 A0A2J7QRF5 O17449 A0A0X9I430 A0A0N0BEN7 D6WSV2 A0A2J7PKK2 A0A067R568 A0A2R7WYX9 V5GSS5 B0G0U7 A0A288SHP6 A0A1B6EWG6 A0A2H8TK23 A0A1W4X4A3 J9K7I9 O76149 A0A0L7LEY4 B1A7Y8 F4WFR2 A0A195B1J3 A0A2A3EFH0 V9IK78 A0A087ZYZ1 A0A158NYQ8 E2AQX7 R4G4U5 A0A026WMP0 D3PFV4 A0A0K2V4C9 A0A1S3DFH4 A0A0A9Z399 A0A023ESD5 T1DI49 A0A1Q3EUF9 U5EUT8 A0A0A9Z013 A0A0A9X553 A0A2J7QSV2 A0A0P6G935 A0A336M9C0 Q17GX9 A0A226F2P2 E0VJE6 A0A1W4VI24 B4LNW8 W8BUE6 D3TR30 B4KME5 A0A1A9YJQ2 A0A0G4AL61 A0A0K8V9H2 A0A1A9UYD7 A0A1B0ARL6 B5DUR1 B4GIN0 B4PB80 A0A0M4EEV8 A0A1I8NLG0 B3MGF8 B4MRR0 B4HPG7 B4J625 B3NJN5 A0A0A1WMS2 A0A1L8EHS1 A0A1A9ZRE2 A0A0K8TT89 A0A1A9WAB8 B4QE31 A0A0L0C7Y7 T1PF10 Q27U48 Q24560 D7RA98 A0A1W4X306 A0A1L8E5M5 R4WCS2 A0A0P6IJV9 A0A224XGD3 A0A1L8E5H8 T1DF12
O01676 A0A2Z5T6R2 I4DMM6 A0A3S2M7U1 A0A1E1W3A9 I4DIS9 Q8T8B2 A9YLN3 A0A212FJY5 F6M9L9 G1E6C4 A0A2J7QIE5 A0A1Y1KNW8 A0A2J7QRF5 O17449 A0A0X9I430 A0A0N0BEN7 D6WSV2 A0A2J7PKK2 A0A067R568 A0A2R7WYX9 V5GSS5 B0G0U7 A0A288SHP6 A0A1B6EWG6 A0A2H8TK23 A0A1W4X4A3 J9K7I9 O76149 A0A0L7LEY4 B1A7Y8 F4WFR2 A0A195B1J3 A0A2A3EFH0 V9IK78 A0A087ZYZ1 A0A158NYQ8 E2AQX7 R4G4U5 A0A026WMP0 D3PFV4 A0A0K2V4C9 A0A1S3DFH4 A0A0A9Z399 A0A023ESD5 T1DI49 A0A1Q3EUF9 U5EUT8 A0A0A9Z013 A0A0A9X553 A0A2J7QSV2 A0A0P6G935 A0A336M9C0 Q17GX9 A0A226F2P2 E0VJE6 A0A1W4VI24 B4LNW8 W8BUE6 D3TR30 B4KME5 A0A1A9YJQ2 A0A0G4AL61 A0A0K8V9H2 A0A1A9UYD7 A0A1B0ARL6 B5DUR1 B4GIN0 B4PB80 A0A0M4EEV8 A0A1I8NLG0 B3MGF8 B4MRR0 B4HPG7 B4J625 B3NJN5 A0A0A1WMS2 A0A1L8EHS1 A0A1A9ZRE2 A0A0K8TT89 A0A1A9WAB8 B4QE31 A0A0L0C7Y7 T1PF10 Q27U48 Q24560 D7RA98 A0A1W4X306 A0A1L8E5M5 R4WCS2 A0A0P6IJV9 A0A224XGD3 A0A1L8E5H8 T1DF12
Pubmed
28756777
22651552
12459208
26354079
19121390
22118469
+ More
28004739 9723895 26826390 18362917 19820115 24845553 26227816 21719571 21347285 20798317 24508170 30249741 25401762 24945155 24330624 17510324 20566863 17994087 24495485 20353571 26252388 15632085 17550304 25830018 26369729 22936249 26108605 25315136 16907828 3119300 10731132 12537572 18327897 22451918 23691247
28004739 9723895 26826390 18362917 19820115 24845553 26227816 21719571 21347285 20798317 24508170 30249741 25401762 24945155 24330624 17510324 20566863 17994087 24495485 20353571 26252388 15632085 17550304 25830018 26369729 22936249 26108605 25315136 16907828 3119300 10731132 12537572 18327897 22451918 23691247
EMBL
RSAL01000012
RVE53453.1
KZ150040
PZC74567.1
AK402356
BAM18978.1
+ More
AB072307 BAB86852.1 KQ459193 KPJ03036.1 GDQN01000168 JAT90886.1 AB003287 BAA19845.1 LC328975 BBA78422.1 AK402544 BAM19166.1 RVE53452.1 GDQN01009594 JAT81460.1 AK401197 BAM17819.1 BABH01035358 AB072308 BAB86853.1 EU234504 ABY47891.1 AGBW02008171 OWR54067.1 JF767013 AEF32528.1 JN029962 AEJ84083.1 NEVH01013629 PNF28355.1 GEZM01080243 JAV62278.1 NEVH01011896 PNF31182.1 AF030547 KU194643 ALP82107.1 KQ435824 KOX72161.1 KQ971352 EFA06663.1 NEVH01024538 PNF16855.1 KK852686 KDR18435.1 KK855763 PTY23825.1 GALX01001232 JAB67234.1 EU373305 ABY66392.1 KX981998 APQ43051.1 GECZ01027490 JAS42279.1 GFXV01002575 MBW14380.1 ABLF02041120 ABLF02041135 AB011069 BAA32102.1 JTDY01001409 KOB73939.1 EU429675 ABZ91904.1 GL888122 EGI66955.1 KQ976681 KYM78155.1 KZ288269 PBC29959.1 JR049240 AEY60906.1 ADTU01004210 ADTU01004211 GL441846 EFN64123.1 ACPB03021733 GAHY01000456 JAA77054.1 KK107151 QOIP01000003 EZA57342.1 RLU24488.1 BT120510 ADD24150.1 HACA01027819 CDW45180.1 GBHO01005781 JAG37823.1 GAPW01001430 JAC12168.1 GALA01001146 JAA93706.1 GFDL01016207 JAV18838.1 GANO01001316 JAB58555.1 GBHO01005780 JAG37824.1 GBHO01028818 JAG14786.1 NEVH01011213 PNF31670.1 GDIQ01047153 GDIQ01002074 JAN47584.1 UFQT01000630 UFQT01000929 SSX25891.1 SSX28043.1 CH477254 EAT45945.1 LNIX01000001 OXA63748.1 DS235222 EEB13502.1 CH940648 EDW61137.1 GAMC01001610 JAC04946.1 EZ423882 ADD20158.1 CH933808 EDW09833.1 KR133399 AKM70869.1 GDHF01016881 JAI35433.1 JXJN01002474 CH674335 EDY71630.1 CH479183 EDW36350.1 CM000158 EDW91493.1 CP012524 ALC42690.1 CH902619 EDV35701.1 CH963850 EDW74799.1 CH480816 EDW48605.1 CH916367 EDW01883.1 CH954179 EDV55213.2 GBXI01014311 JAC99980.1 GFDG01000517 JAV18282.1 GDAI01000029 JAI17574.1 CM000362 CM002911 EDX07818.1 KMY95122.1 JRES01001535 JRES01000890 KNC22195.1 KNC27524.1 KA647387 AFP62016.1 DQ377071 M20419 AE013599 BT003242 HM047134 ADI24738.1 GFDF01000155 JAV13929.1 AK416954 BAN20169.1 GDIQ01040660 GDIQ01003233 JAN91504.1 GFTR01006332 JAW10094.1 GFDF01000126 JAV13958.1 GAMD01003291 JAA98299.1
AB072307 BAB86852.1 KQ459193 KPJ03036.1 GDQN01000168 JAT90886.1 AB003287 BAA19845.1 LC328975 BBA78422.1 AK402544 BAM19166.1 RVE53452.1 GDQN01009594 JAT81460.1 AK401197 BAM17819.1 BABH01035358 AB072308 BAB86853.1 EU234504 ABY47891.1 AGBW02008171 OWR54067.1 JF767013 AEF32528.1 JN029962 AEJ84083.1 NEVH01013629 PNF28355.1 GEZM01080243 JAV62278.1 NEVH01011896 PNF31182.1 AF030547 KU194643 ALP82107.1 KQ435824 KOX72161.1 KQ971352 EFA06663.1 NEVH01024538 PNF16855.1 KK852686 KDR18435.1 KK855763 PTY23825.1 GALX01001232 JAB67234.1 EU373305 ABY66392.1 KX981998 APQ43051.1 GECZ01027490 JAS42279.1 GFXV01002575 MBW14380.1 ABLF02041120 ABLF02041135 AB011069 BAA32102.1 JTDY01001409 KOB73939.1 EU429675 ABZ91904.1 GL888122 EGI66955.1 KQ976681 KYM78155.1 KZ288269 PBC29959.1 JR049240 AEY60906.1 ADTU01004210 ADTU01004211 GL441846 EFN64123.1 ACPB03021733 GAHY01000456 JAA77054.1 KK107151 QOIP01000003 EZA57342.1 RLU24488.1 BT120510 ADD24150.1 HACA01027819 CDW45180.1 GBHO01005781 JAG37823.1 GAPW01001430 JAC12168.1 GALA01001146 JAA93706.1 GFDL01016207 JAV18838.1 GANO01001316 JAB58555.1 GBHO01005780 JAG37824.1 GBHO01028818 JAG14786.1 NEVH01011213 PNF31670.1 GDIQ01047153 GDIQ01002074 JAN47584.1 UFQT01000630 UFQT01000929 SSX25891.1 SSX28043.1 CH477254 EAT45945.1 LNIX01000001 OXA63748.1 DS235222 EEB13502.1 CH940648 EDW61137.1 GAMC01001610 JAC04946.1 EZ423882 ADD20158.1 CH933808 EDW09833.1 KR133399 AKM70869.1 GDHF01016881 JAI35433.1 JXJN01002474 CH674335 EDY71630.1 CH479183 EDW36350.1 CM000158 EDW91493.1 CP012524 ALC42690.1 CH902619 EDV35701.1 CH963850 EDW74799.1 CH480816 EDW48605.1 CH916367 EDW01883.1 CH954179 EDV55213.2 GBXI01014311 JAC99980.1 GFDG01000517 JAV18282.1 GDAI01000029 JAI17574.1 CM000362 CM002911 EDX07818.1 KMY95122.1 JRES01001535 JRES01000890 KNC22195.1 KNC27524.1 KA647387 AFP62016.1 DQ377071 M20419 AE013599 BT003242 HM047134 ADI24738.1 GFDF01000155 JAV13929.1 AK416954 BAN20169.1 GDIQ01040660 GDIQ01003233 JAN91504.1 GFTR01006332 JAW10094.1 GFDF01000126 JAV13958.1 GAMD01003291 JAA98299.1
Proteomes
UP000283053
UP000053268
UP000005204
UP000007151
UP000235965
UP000053105
+ More
UP000007266 UP000027135 UP000192223 UP000007819 UP000037510 UP000007755 UP000078540 UP000242457 UP000005203 UP000005205 UP000000311 UP000015103 UP000053097 UP000279307 UP000079169 UP000008820 UP000198287 UP000009046 UP000192221 UP000008792 UP000009192 UP000092443 UP000078200 UP000092460 UP000001819 UP000008744 UP000002282 UP000092553 UP000095300 UP000007801 UP000007798 UP000001292 UP000001070 UP000008711 UP000092445 UP000091820 UP000000304 UP000037069 UP000095301 UP000092444 UP000000803
UP000007266 UP000027135 UP000192223 UP000007819 UP000037510 UP000007755 UP000078540 UP000242457 UP000005203 UP000005205 UP000000311 UP000015103 UP000053097 UP000279307 UP000079169 UP000008820 UP000198287 UP000009046 UP000192221 UP000008792 UP000009192 UP000092443 UP000078200 UP000092460 UP000001819 UP000008744 UP000002282 UP000092553 UP000095300 UP000007801 UP000007798 UP000001292 UP000001070 UP000008711 UP000092445 UP000091820 UP000000304 UP000037069 UP000095301 UP000092444 UP000000803
Interpro
IPR023123
Tubulin_C
+ More
IPR000217 Tubulin
IPR018316 Tubulin/FtsZ_2-layer-sand-dom
IPR036525 Tubulin/FtsZ_GTPase_sf
IPR017975 Tubulin_CS
IPR003008 Tubulin_FtsZ_GTPase
IPR008280 Tub_FtsZ_C
IPR013838 Beta-tubulin_BS
IPR002453 Beta_tubulin
IPR037103 Tubulin/FtsZ_C_sf
IPR019410 Methyltransf_16
IPR029063 SAM-dependent_MTases
IPR000217 Tubulin
IPR018316 Tubulin/FtsZ_2-layer-sand-dom
IPR036525 Tubulin/FtsZ_GTPase_sf
IPR017975 Tubulin_CS
IPR003008 Tubulin_FtsZ_GTPase
IPR008280 Tub_FtsZ_C
IPR013838 Beta-tubulin_BS
IPR002453 Beta_tubulin
IPR037103 Tubulin/FtsZ_C_sf
IPR019410 Methyltransf_16
IPR029063 SAM-dependent_MTases
Gene 3D
ProteinModelPortal
A0A3S2NZY9
A0A2W1BMR7
I4DM38
Q8T8B3
A0A194QHV4
A0A1E1WVR1
+ More
O01676 A0A2Z5T6R2 I4DMM6 A0A3S2M7U1 A0A1E1W3A9 I4DIS9 Q8T8B2 A9YLN3 A0A212FJY5 F6M9L9 G1E6C4 A0A2J7QIE5 A0A1Y1KNW8 A0A2J7QRF5 O17449 A0A0X9I430 A0A0N0BEN7 D6WSV2 A0A2J7PKK2 A0A067R568 A0A2R7WYX9 V5GSS5 B0G0U7 A0A288SHP6 A0A1B6EWG6 A0A2H8TK23 A0A1W4X4A3 J9K7I9 O76149 A0A0L7LEY4 B1A7Y8 F4WFR2 A0A195B1J3 A0A2A3EFH0 V9IK78 A0A087ZYZ1 A0A158NYQ8 E2AQX7 R4G4U5 A0A026WMP0 D3PFV4 A0A0K2V4C9 A0A1S3DFH4 A0A0A9Z399 A0A023ESD5 T1DI49 A0A1Q3EUF9 U5EUT8 A0A0A9Z013 A0A0A9X553 A0A2J7QSV2 A0A0P6G935 A0A336M9C0 Q17GX9 A0A226F2P2 E0VJE6 A0A1W4VI24 B4LNW8 W8BUE6 D3TR30 B4KME5 A0A1A9YJQ2 A0A0G4AL61 A0A0K8V9H2 A0A1A9UYD7 A0A1B0ARL6 B5DUR1 B4GIN0 B4PB80 A0A0M4EEV8 A0A1I8NLG0 B3MGF8 B4MRR0 B4HPG7 B4J625 B3NJN5 A0A0A1WMS2 A0A1L8EHS1 A0A1A9ZRE2 A0A0K8TT89 A0A1A9WAB8 B4QE31 A0A0L0C7Y7 T1PF10 Q27U48 Q24560 D7RA98 A0A1W4X306 A0A1L8E5M5 R4WCS2 A0A0P6IJV9 A0A224XGD3 A0A1L8E5H8 T1DF12
O01676 A0A2Z5T6R2 I4DMM6 A0A3S2M7U1 A0A1E1W3A9 I4DIS9 Q8T8B2 A9YLN3 A0A212FJY5 F6M9L9 G1E6C4 A0A2J7QIE5 A0A1Y1KNW8 A0A2J7QRF5 O17449 A0A0X9I430 A0A0N0BEN7 D6WSV2 A0A2J7PKK2 A0A067R568 A0A2R7WYX9 V5GSS5 B0G0U7 A0A288SHP6 A0A1B6EWG6 A0A2H8TK23 A0A1W4X4A3 J9K7I9 O76149 A0A0L7LEY4 B1A7Y8 F4WFR2 A0A195B1J3 A0A2A3EFH0 V9IK78 A0A087ZYZ1 A0A158NYQ8 E2AQX7 R4G4U5 A0A026WMP0 D3PFV4 A0A0K2V4C9 A0A1S3DFH4 A0A0A9Z399 A0A023ESD5 T1DI49 A0A1Q3EUF9 U5EUT8 A0A0A9Z013 A0A0A9X553 A0A2J7QSV2 A0A0P6G935 A0A336M9C0 Q17GX9 A0A226F2P2 E0VJE6 A0A1W4VI24 B4LNW8 W8BUE6 D3TR30 B4KME5 A0A1A9YJQ2 A0A0G4AL61 A0A0K8V9H2 A0A1A9UYD7 A0A1B0ARL6 B5DUR1 B4GIN0 B4PB80 A0A0M4EEV8 A0A1I8NLG0 B3MGF8 B4MRR0 B4HPG7 B4J625 B3NJN5 A0A0A1WMS2 A0A1L8EHS1 A0A1A9ZRE2 A0A0K8TT89 A0A1A9WAB8 B4QE31 A0A0L0C7Y7 T1PF10 Q27U48 Q24560 D7RA98 A0A1W4X306 A0A1L8E5M5 R4WCS2 A0A0P6IJV9 A0A224XGD3 A0A1L8E5H8 T1DF12
PDB
5Z4U
E-value=0,
Score=2174
Ontologies
GO
PANTHER
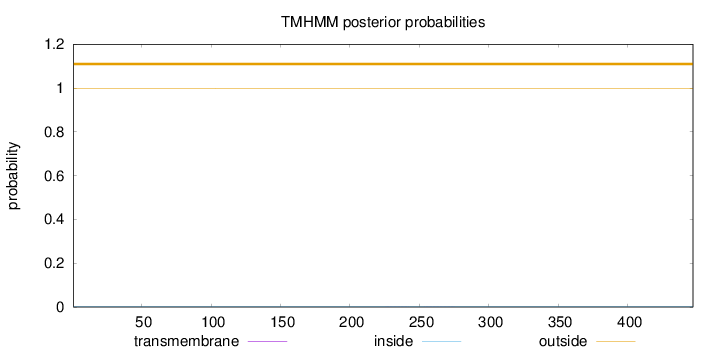
Topology
Subcellular location
Length:
447
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01066
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00368
outside
1 - 447
Population Genetic Test Statistics
Pi
23.615503
Theta
22.82971
Tajima's D
0.105176
CLR
0.11307
CSRT
0.401429928503575
Interpretation
Uncertain
