Gene
KWMTBOMO12284
Pre Gene Modal
BGIBMGA009154
Annotation
uridine_5'-monophosphate_synthase_[Danaus_plexippus]
Full name
Orotate phosphoribosyltransferase
Location in the cell
Cytoplasmic Reliability : 2.275
Sequence
CDS
ATGACGTCCACTCTACAGGATCTGGCAGTGCAGTTATTCGACGCAGGAGCAGTTCGACTTGAAGACATAGAGGCGAAGGTCGGCAGGCGAACCCCCATCTACTTCGACCTACGAGTCATCGTTTCGCACCCTAAGCTGATGGTAGCAGTGTCACATCAATTGGAGAAACTCATTAAAGACGTTCATCACGATATGATATGTGGAGTTCCATACGCTGCACTGCCTCTTGCAGTAGTCATCTCCATCAACACGGACAGACCAATGATCATGAAGAGAAAAGAAACAAAATTATATGCCACAAAGAAATTACTGGAGGGCGTGTATCAAAAAAATCAGACGTGCCTTGTTGTTGAAGATGTAGTGACATCTGGCGGCAGCTTGCTAGAAACAGTTGAAACTTTACGCAAAGAAGGTTTGATAGTTACACATGCCGTAATAGTACTCGAGCGGGAACAAGGTGGCGCTAGTGTACTCGAAGCGAATGGTATTACAGTGAAATCACTTTTCACATTAACTAATTTAGTGAGAATGCTTCATAAAGCGGGTAGAATTGACAATGACACAGTCGAATTAATATTTGATCATATAAAGGAATGCCAATTTGGAACGAAAGATAATTTGGAGTAG
Protein
MTSTLQDLAVQLFDAGAVRLEDIEAKVGRRTPIYFDLRVIVSHPKLMVAVSHQLEKLIKDVHHDMICGVPYAALPLAVVISINTDRPMIMKRKETKLYATKKLLEGVYQKNQTCLVVEDVVTSGGSLLETVETLRKEGLIVTHAVIVLEREQGGASVLEANGITVKSLFTLTNLVRMLHKAGRIDNDTVELIFDHIKECQFGTKDNLE
Summary
Description
Catalyzes the transfer of a ribosyl phosphate group from 5-phosphoribose 1-diphosphate to orotate, leading to the formation of orotidine monophosphate (OMP).
Catalytic Activity
diphosphate + orotidine 5'-phosphate = 5-phospho-alpha-D-ribose 1-diphosphate + orotate
Cofactor
Mg(2+)
Subunit
Homodimer.
Similarity
Belongs to the purine/pyrimidine phosphoribosyltransferase family. PyrE subfamily.
Uniprot
H9JI06
A0A212FK36
A0A2H1X1H1
A0A2A4IZA9
A0A0N0PB06
A0A2W1BI85
+ More
A0A3S2NKS2 A0A194QBV5 A0A0L7LEW0 A0A2H1V5H1 A0A2A4JS88 A0A1B6DH00 E2AMC4 S4NGX6 A0A182SJA8 A0A2M3Z341 H9J7Y6 T1DQ66 A0A1B6KY40 Q2F642 W5JU62 A0A2M4BL31 A0A2M4BL32 A0A2M4AG61 A0A336KMV5 A0A1B6K1V9 A0A1I8QBG3 K7IX88 A0A232FC27 A0A182IR66 A0A026VYJ3 B0W331 A0A182YGP3 E9IMV3 A0A023F7U3 A0A1B6EQX9 A0A1B6G5N9 A0A182QCX4 A0A3B3R2R5 A0A023ES01 A0A195D1G3 U5EU74 A0A151XDF2 F4X083 A0A0L0CLW6 A0A1Q3F1P9 A0A182FSD4 A0A182JTD1 A0A0L7LET4 Q16QF3 A0A182UWE0 A0A1S4FSU7 A0A182U319 Q7QB57 A0A194QGS7 A0A158NHC2 A0A0D2V1M4 A0A195AZK8 B4JSX4 A0A0G1VN04 A0A182HBI2 A0A224XP35 A0A182RYP4 N6ST75 A0A182LTP8 A0A232EWX5 E9J1A5 A0A0D2U139 A0A1U8J468 A0A0V0GAW1 A0A1L8DY49 A0A182P6S0 A0A182NLC8 R7U604 A0A340TBA6 Q7ZVR7 A0A0P4VL83 R4FNI1 B4M4V8 A0A0M8ZX99 A0A182XJ45 A0A182I7R6 A0A154PPC3 A0A397YLM6 A0A1F6LJI1 A0A182L7A8 A0A146P8A5 A0A0C9QBM4 B4KE16 A0A1U8NVE8 A0A061F815 A0A061F719 A0A146MP24 B4NH48 A0A0B0MB88 A0A2J6JYY7 A0A3P6BSA4 A0A084VDC6 A0A1I8ND48 T1PCL7
A0A3S2NKS2 A0A194QBV5 A0A0L7LEW0 A0A2H1V5H1 A0A2A4JS88 A0A1B6DH00 E2AMC4 S4NGX6 A0A182SJA8 A0A2M3Z341 H9J7Y6 T1DQ66 A0A1B6KY40 Q2F642 W5JU62 A0A2M4BL31 A0A2M4BL32 A0A2M4AG61 A0A336KMV5 A0A1B6K1V9 A0A1I8QBG3 K7IX88 A0A232FC27 A0A182IR66 A0A026VYJ3 B0W331 A0A182YGP3 E9IMV3 A0A023F7U3 A0A1B6EQX9 A0A1B6G5N9 A0A182QCX4 A0A3B3R2R5 A0A023ES01 A0A195D1G3 U5EU74 A0A151XDF2 F4X083 A0A0L0CLW6 A0A1Q3F1P9 A0A182FSD4 A0A182JTD1 A0A0L7LET4 Q16QF3 A0A182UWE0 A0A1S4FSU7 A0A182U319 Q7QB57 A0A194QGS7 A0A158NHC2 A0A0D2V1M4 A0A195AZK8 B4JSX4 A0A0G1VN04 A0A182HBI2 A0A224XP35 A0A182RYP4 N6ST75 A0A182LTP8 A0A232EWX5 E9J1A5 A0A0D2U139 A0A1U8J468 A0A0V0GAW1 A0A1L8DY49 A0A182P6S0 A0A182NLC8 R7U604 A0A340TBA6 Q7ZVR7 A0A0P4VL83 R4FNI1 B4M4V8 A0A0M8ZX99 A0A182XJ45 A0A182I7R6 A0A154PPC3 A0A397YLM6 A0A1F6LJI1 A0A182L7A8 A0A146P8A5 A0A0C9QBM4 B4KE16 A0A1U8NVE8 A0A061F815 A0A061F719 A0A146MP24 B4NH48 A0A0B0MB88 A0A2J6JYY7 A0A3P6BSA4 A0A084VDC6 A0A1I8ND48 T1PCL7
EC Number
2.4.2.10
Pubmed
19121390
22118469
26354079
28756777
26227816
20798317
+ More
23622113 20920257 23761445 20075255 28648823 24508170 30249741 25244985 21282665 25474469 29240929 24945155 21719571 26108605 17510324 12364791 14747013 17210077 21347285 23257886 17994087 26483478 23537049 25893780 23254933 27129103 27774985 20966253 23731509 28401891 24438588 25315136
23622113 20920257 23761445 20075255 28648823 24508170 30249741 25244985 21282665 25474469 29240929 24945155 21719571 26108605 17510324 12364791 14747013 17210077 21347285 23257886 17994087 26483478 23537049 25893780 23254933 27129103 27774985 20966253 23731509 28401891 24438588 25315136
EMBL
BABH01035357
AGBW02008171
OWR54069.1
ODYU01012659
SOQ59088.1
NWSH01004290
+ More
PCG65267.1 KQ461072 KPJ09818.1 KZ150040 PZC74568.1 RSAL01000012 RVE53451.1 KQ459193 KPJ03038.1 JTDY01001409 KOB73940.1 ODYU01000778 SOQ36083.1 NWSH01000703 PCG74689.1 GEDC01012344 JAS24954.1 GL440811 EFN65415.1 GAIX01014549 JAA78011.1 GGFM01002107 MBW22858.1 BABH01020166 GAMD01002719 JAA98871.1 GEBQ01023656 JAT16321.1 DQ311230 ABD36175.1 ADMH02000361 ETN66833.1 GGFJ01004639 MBW53780.1 GGFJ01004638 MBW53779.1 GGFK01006452 MBW39773.1 UFQS01000645 UFQT01000410 UFQT01000645 SSX05697.1 SSX24054.1 GECU01002298 JAT05409.1 AAZX01000583 NNAY01000465 OXU28182.1 KK107566 QOIP01000006 EZA48873.1 RLU21950.1 DS231830 EDS30739.1 GL764255 EFZ18097.1 GBBI01001433 JAC17279.1 GECZ01029408 JAS40361.1 GECZ01012037 JAS57732.1 AXCN02000121 GAPW01001603 JAC11995.1 KQ976973 KYN06707.1 GANO01001613 JAB58258.1 KQ982294 KYQ58387.1 GL888493 EGI60148.1 JRES01000211 KNC33256.1 GFDL01013586 JAV21459.1 JTDY01001365 KOB74068.1 CH477750 EAT36623.1 AAAB01008880 EAA08525.3 KQ459232 KPJ02651.1 ADTU01015566 CM001748 KJB62735.1 KQ976698 KYM77470.1 CH916373 EDV94864.1 LCPZ01000022 KKW07853.1 JXUM01032039 JXUM01032040 KQ560942 KXJ80255.1 GFTR01006503 JAW09923.1 APGK01057391 KB741280 KB631604 ENN70894.1 ERL84613.1 AXCM01003761 NNAY01001812 OXU22847.1 GL767586 EFZ13419.1 KJB62734.1 GECL01000923 JAP05201.1 GFDF01002899 JAV11185.1 AMQN01001733 KB305005 ELU01511.1 BC045440 BC055000 BC164016 AAH45440.1 AAH55000.1 AAI64016.1 GDKW01001438 JAI55157.1 ACPB03010572 GAHY01001284 JAA76226.1 CH940652 EDW59669.1 KQ435837 KOX71473.1 APCN01002549 KQ435010 KZC13745.1 CM010634 RID54301.1 MFPS01000007 OGH59499.1 GCES01146131 JAQ40191.1 GBYB01011813 JAG81580.1 CH933806 EDW16037.1 CM001885 EOY12847.1 EOY12846.1 GCES01165942 JAQ20380.1 CH964272 EDW84545.1 JRRC01025018 KHF98024.1 NBSK01009389 PLY68034.1 LR031574 VDC99181.1 ATLV01011254 KE524659 KFB35970.1 KA645885 AFP60514.1
PCG65267.1 KQ461072 KPJ09818.1 KZ150040 PZC74568.1 RSAL01000012 RVE53451.1 KQ459193 KPJ03038.1 JTDY01001409 KOB73940.1 ODYU01000778 SOQ36083.1 NWSH01000703 PCG74689.1 GEDC01012344 JAS24954.1 GL440811 EFN65415.1 GAIX01014549 JAA78011.1 GGFM01002107 MBW22858.1 BABH01020166 GAMD01002719 JAA98871.1 GEBQ01023656 JAT16321.1 DQ311230 ABD36175.1 ADMH02000361 ETN66833.1 GGFJ01004639 MBW53780.1 GGFJ01004638 MBW53779.1 GGFK01006452 MBW39773.1 UFQS01000645 UFQT01000410 UFQT01000645 SSX05697.1 SSX24054.1 GECU01002298 JAT05409.1 AAZX01000583 NNAY01000465 OXU28182.1 KK107566 QOIP01000006 EZA48873.1 RLU21950.1 DS231830 EDS30739.1 GL764255 EFZ18097.1 GBBI01001433 JAC17279.1 GECZ01029408 JAS40361.1 GECZ01012037 JAS57732.1 AXCN02000121 GAPW01001603 JAC11995.1 KQ976973 KYN06707.1 GANO01001613 JAB58258.1 KQ982294 KYQ58387.1 GL888493 EGI60148.1 JRES01000211 KNC33256.1 GFDL01013586 JAV21459.1 JTDY01001365 KOB74068.1 CH477750 EAT36623.1 AAAB01008880 EAA08525.3 KQ459232 KPJ02651.1 ADTU01015566 CM001748 KJB62735.1 KQ976698 KYM77470.1 CH916373 EDV94864.1 LCPZ01000022 KKW07853.1 JXUM01032039 JXUM01032040 KQ560942 KXJ80255.1 GFTR01006503 JAW09923.1 APGK01057391 KB741280 KB631604 ENN70894.1 ERL84613.1 AXCM01003761 NNAY01001812 OXU22847.1 GL767586 EFZ13419.1 KJB62734.1 GECL01000923 JAP05201.1 GFDF01002899 JAV11185.1 AMQN01001733 KB305005 ELU01511.1 BC045440 BC055000 BC164016 AAH45440.1 AAH55000.1 AAI64016.1 GDKW01001438 JAI55157.1 ACPB03010572 GAHY01001284 JAA76226.1 CH940652 EDW59669.1 KQ435837 KOX71473.1 APCN01002549 KQ435010 KZC13745.1 CM010634 RID54301.1 MFPS01000007 OGH59499.1 GCES01146131 JAQ40191.1 GBYB01011813 JAG81580.1 CH933806 EDW16037.1 CM001885 EOY12847.1 EOY12846.1 GCES01165942 JAQ20380.1 CH964272 EDW84545.1 JRRC01025018 KHF98024.1 NBSK01009389 PLY68034.1 LR031574 VDC99181.1 ATLV01011254 KE524659 KFB35970.1 KA645885 AFP60514.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000283053
UP000053268
+ More
UP000037510 UP000000311 UP000075901 UP000000673 UP000095300 UP000002358 UP000215335 UP000075880 UP000053097 UP000279307 UP000002320 UP000076408 UP000075886 UP000261540 UP000078542 UP000075809 UP000007755 UP000037069 UP000069272 UP000075881 UP000008820 UP000075903 UP000075902 UP000007062 UP000005205 UP000032304 UP000078540 UP000001070 UP000069940 UP000249989 UP000075900 UP000019118 UP000030742 UP000075883 UP000189702 UP000075885 UP000075884 UP000014760 UP000075920 UP000015103 UP000008792 UP000053105 UP000076407 UP000075840 UP000076502 UP000075882 UP000009192 UP000026915 UP000007798 UP000235145 UP000030765 UP000095301
UP000037510 UP000000311 UP000075901 UP000000673 UP000095300 UP000002358 UP000215335 UP000075880 UP000053097 UP000279307 UP000002320 UP000076408 UP000075886 UP000261540 UP000078542 UP000075809 UP000007755 UP000037069 UP000069272 UP000075881 UP000008820 UP000075903 UP000075902 UP000007062 UP000005205 UP000032304 UP000078540 UP000001070 UP000069940 UP000249989 UP000075900 UP000019118 UP000030742 UP000075883 UP000189702 UP000075885 UP000075884 UP000014760 UP000075920 UP000015103 UP000008792 UP000053105 UP000076407 UP000075840 UP000076502 UP000075882 UP000009192 UP000026915 UP000007798 UP000235145 UP000030765 UP000095301
Pfam
Interpro
IPR000836
PRibTrfase_dom
+ More
IPR029057 PRTase-like
IPR004467 Or_phspho_trans_dom
IPR023031 OPRT
IPR018089 OMPdecase_AS
IPR001754 OMPdeCOase_dom
IPR013785 Aldolase_TIM
IPR011060 RibuloseP-bd_barrel
IPR014732 OMPdecase
IPR033523 IRE2
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR010513 KEN_dom
IPR018997 PUB_domain
IPR018391 PQQ_beta_propeller_repeat
IPR038357 KEN_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR000719 Prot_kinase_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036372 BEACH_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR036322 WD40_repeat_dom_sf
IPR000409 BEACH_dom
IPR002877 rRNA_MeTrfase_FtsJ_dom
IPR015507 rRNA-MeTfrase_E
IPR029063 SAM-dependent_MTases
IPR017441 Protein_kinase_ATP_BS
IPR029057 PRTase-like
IPR004467 Or_phspho_trans_dom
IPR023031 OPRT
IPR018089 OMPdecase_AS
IPR001754 OMPdeCOase_dom
IPR013785 Aldolase_TIM
IPR011060 RibuloseP-bd_barrel
IPR014732 OMPdecase
IPR033523 IRE2
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR010513 KEN_dom
IPR018997 PUB_domain
IPR018391 PQQ_beta_propeller_repeat
IPR038357 KEN_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR000719 Prot_kinase_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036372 BEACH_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR036322 WD40_repeat_dom_sf
IPR000409 BEACH_dom
IPR002877 rRNA_MeTrfase_FtsJ_dom
IPR015507 rRNA-MeTfrase_E
IPR029063 SAM-dependent_MTases
IPR017441 Protein_kinase_ATP_BS
SUPFAM
ProteinModelPortal
H9JI06
A0A212FK36
A0A2H1X1H1
A0A2A4IZA9
A0A0N0PB06
A0A2W1BI85
+ More
A0A3S2NKS2 A0A194QBV5 A0A0L7LEW0 A0A2H1V5H1 A0A2A4JS88 A0A1B6DH00 E2AMC4 S4NGX6 A0A182SJA8 A0A2M3Z341 H9J7Y6 T1DQ66 A0A1B6KY40 Q2F642 W5JU62 A0A2M4BL31 A0A2M4BL32 A0A2M4AG61 A0A336KMV5 A0A1B6K1V9 A0A1I8QBG3 K7IX88 A0A232FC27 A0A182IR66 A0A026VYJ3 B0W331 A0A182YGP3 E9IMV3 A0A023F7U3 A0A1B6EQX9 A0A1B6G5N9 A0A182QCX4 A0A3B3R2R5 A0A023ES01 A0A195D1G3 U5EU74 A0A151XDF2 F4X083 A0A0L0CLW6 A0A1Q3F1P9 A0A182FSD4 A0A182JTD1 A0A0L7LET4 Q16QF3 A0A182UWE0 A0A1S4FSU7 A0A182U319 Q7QB57 A0A194QGS7 A0A158NHC2 A0A0D2V1M4 A0A195AZK8 B4JSX4 A0A0G1VN04 A0A182HBI2 A0A224XP35 A0A182RYP4 N6ST75 A0A182LTP8 A0A232EWX5 E9J1A5 A0A0D2U139 A0A1U8J468 A0A0V0GAW1 A0A1L8DY49 A0A182P6S0 A0A182NLC8 R7U604 A0A340TBA6 Q7ZVR7 A0A0P4VL83 R4FNI1 B4M4V8 A0A0M8ZX99 A0A182XJ45 A0A182I7R6 A0A154PPC3 A0A397YLM6 A0A1F6LJI1 A0A182L7A8 A0A146P8A5 A0A0C9QBM4 B4KE16 A0A1U8NVE8 A0A061F815 A0A061F719 A0A146MP24 B4NH48 A0A0B0MB88 A0A2J6JYY7 A0A3P6BSA4 A0A084VDC6 A0A1I8ND48 T1PCL7
A0A3S2NKS2 A0A194QBV5 A0A0L7LEW0 A0A2H1V5H1 A0A2A4JS88 A0A1B6DH00 E2AMC4 S4NGX6 A0A182SJA8 A0A2M3Z341 H9J7Y6 T1DQ66 A0A1B6KY40 Q2F642 W5JU62 A0A2M4BL31 A0A2M4BL32 A0A2M4AG61 A0A336KMV5 A0A1B6K1V9 A0A1I8QBG3 K7IX88 A0A232FC27 A0A182IR66 A0A026VYJ3 B0W331 A0A182YGP3 E9IMV3 A0A023F7U3 A0A1B6EQX9 A0A1B6G5N9 A0A182QCX4 A0A3B3R2R5 A0A023ES01 A0A195D1G3 U5EU74 A0A151XDF2 F4X083 A0A0L0CLW6 A0A1Q3F1P9 A0A182FSD4 A0A182JTD1 A0A0L7LET4 Q16QF3 A0A182UWE0 A0A1S4FSU7 A0A182U319 Q7QB57 A0A194QGS7 A0A158NHC2 A0A0D2V1M4 A0A195AZK8 B4JSX4 A0A0G1VN04 A0A182HBI2 A0A224XP35 A0A182RYP4 N6ST75 A0A182LTP8 A0A232EWX5 E9J1A5 A0A0D2U139 A0A1U8J468 A0A0V0GAW1 A0A1L8DY49 A0A182P6S0 A0A182NLC8 R7U604 A0A340TBA6 Q7ZVR7 A0A0P4VL83 R4FNI1 B4M4V8 A0A0M8ZX99 A0A182XJ45 A0A182I7R6 A0A154PPC3 A0A397YLM6 A0A1F6LJI1 A0A182L7A8 A0A146P8A5 A0A0C9QBM4 B4KE16 A0A1U8NVE8 A0A061F815 A0A061F719 A0A146MP24 B4NH48 A0A0B0MB88 A0A2J6JYY7 A0A3P6BSA4 A0A084VDC6 A0A1I8ND48 T1PCL7
PDB
2WNS
E-value=4.30504e-41,
Score=419
Ontologies
PATHWAY
GO
PANTHER
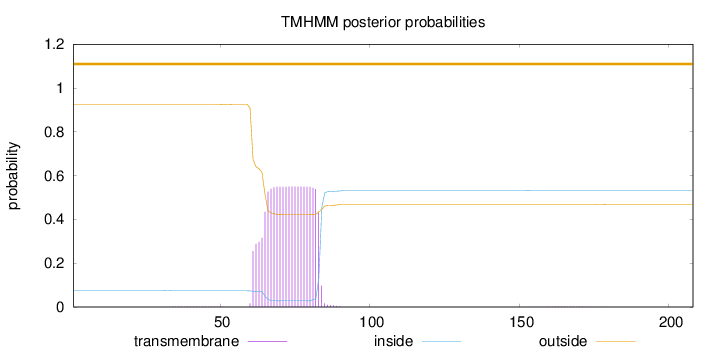
Topology
Length:
208
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.5397
Exp number, first 60 AAs:
0.05361
Total prob of N-in:
0.07582
outside
1 - 208
Population Genetic Test Statistics
Pi
24.06874
Theta
20.758053
Tajima's D
0.438025
CLR
1.458758
CSRT
0.500224988750562
Interpretation
Uncertain
