Gene
KWMTBOMO12282 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009132
Annotation
beta-tubulin_[Bombyx_mori]
Full name
Tubulin beta chain
+ More
Tubulin beta-3 chain
Tubulin beta-3 chain
Alternative Name
Beta-3-tubulin
Location in the cell
Cytoplasmic Reliability : 3.44
Sequence
CDS
ATGTTCTGGGAGATAATATCGGAAGAGCACGGCATCGACCCTACCGGCGTCTACCGCGGCACCAGCGACCTCCAGCTCGAGAGGATCTCTGTTTACTATAATGAGGCCTCTGTTGCGACGGCGGAGAGCGGCGGGAAGTATGTCCCCCGCGCCATCCTCCTCGACCTCGAGCCCGGCACCATGGACGCCGTGCGCTCCGGCGCCTACGGGCAACTCTTCCGGCCGGACAACTTCGTCTTCGGCCAGTCCGGCGCCGGCAACAACTGGGCCAAGGGACACTACACGGAGGGCGCTGAGCTCGTCGACGCCGTCCTTGATGTAGTACGCAAGGAGTGTGAGAACTGCGATTGCCTCCAGGGTTTCCAACTGACTCATTCCCTCGGCGGCGGCACCGGTTCCGGCATGGGCACCCTCCTCATCTCAAAGATCCGTGAAGAGTACCCCGACAGAATCATGAACACATACTCAGTAGTCCCCTCGCCCAAAGTATCAGACACCGTCGTCGAACCATACAACGCAGTTCTCTCCATCCATCAACTAGTCGAGAATACAGACGAAACTTACTGCATAGACAATGAGGCCCTTTACGATATCTGCTACAGGACTCTCAAGGTACCGAACCCAACGTACGGCGACCTGAACCACCTGGTGTCGCTGACCATGTCCGGCGTGACGACGTGCCTGCGGTTCCCCGGACAGCTGAACGCCGACCTGCGCAAGCTGGCGGTCAACATGGTGCCCTTCCCGCGTCTGCACTTCTTCATGCCCGGCTTCGCGCCGCTGACGTCACGCGGCAGCCAGCAGTACCGCGCGCTCACCGTGCCGGAGCTCACGCAGCAGATGTTCGACGCCAAGAACATGATGGCCGCCTGCGACCCGCGCCACGGCCGCTACCTCACCGTCGCCGCCATCTTCCGCGGCCGCATGTCCATGAAGGAGGTCGACGAGCAGATGCTCTCCATCCAAAACAAGAACAGCAGCTTCTTCGTCGAGTGGATCCCCAACAACGTGAAAACAGCCGTGTGCGACATCCCTCCCAAGGGTCTGAAGATGTCGTCTACGTTTATCGGGAACACGACAGCCATCCAGGAACTATTCAAGAGAATATCCGAGCAGTTCTCTGCCATGTTCAGACGTAAAGCTTTCTTGCACTGGTACACCGGCGAGGGCATGGACGAGATGGAGTTCAACGAAGCAGAGAGCAACGTCAACGATCTGGTGTCTGAGTACCAACAGTACCAGGAGGCCACCGCGGAAGACGACACCGAGTTCGACCAAGAAGACCTGGAGGAGTTGGCGCAGGACGAGCACCATGACTAG
Protein
MFWEIISEEHGIDPTGVYRGTSDLQLERISVYYNEASVATAESGGKYVPRAILLDLEPGTMDAVRSGAYGQLFRPDNFVFGQSGAGNNWAKGHYTEGAELVDAVLDVVRKECENCDCLQGFQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTYSVVPSPKVSDTVVEPYNAVLSIHQLVENTDETYCIDNEALYDICYRTLKVPNPTYGDLNHLVSLTMSGVTTCLRFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLTVAAIFRGRMSMKEVDEQMLSIQNKNSSFFVEWIPNNVKTAVCDIPPKGLKMSSTFIGNTTAIQELFKRISEQFSAMFRRKAFLHWYTGEGMDEMEFNEAESNVNDLVSEYQQYQEATAEDDTEFDQEDLEELAQDEHHD
Summary
Description
Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
Subunit
Dimer of alpha and beta chains. A typical microtubule is a hollow water-filled tube with an outer diameter of 25 nm and an inner diameter of 15 nM. Alpha-beta heterodimers associate head-to-tail to form protofilaments running lengthwise along the microtubule wall with the beta-tubulin subunit facing the microtubule plus end conferring a structural polarity. Microtubules usually have 13 protofilaments but different protofilament numbers can be found in some organisms and specialized cells.
Miscellaneous
Beta-3 is a developmentally regulated isoform.
Similarity
Belongs to the tubulin family.
Keywords
Complete proteome
Cytoplasm
Cytoskeleton
GTP-binding
Microtubule
Nucleotide-binding
Reference proteome
Feature
chain Tubulin beta-3 chain
Uniprot
A0A2H1WBT4
A0A2W1BHJ4
Q8T8B1
A0A212F8H2
I4DJ84
A0A0N1INH4
+ More
A0A2P8YDV3 A0A182H8I8 A0A1L8E625 A0A1B0C8Q4 A0A182UPL1 A0A182TJQ3 Q17GX8 A0A1S4F2Y4 A0A023EUS0 A0A182QZM6 B0W7F0 A0A182MY89 A0A182MT22 A0A182WUV4 A0A182HYE9 A0A084WAZ5 Q7PPR6 A0A182PJ94 A0A182YF86 A0A182K7P5 A0A182LPX7 A0A182VT65 T1DPP7 A0A182RIT5 A0A182JB51 A0A0L7L7X9 A0A310SK65 A0A1B0G7V4 A0A026WMT3 A0A0L7RDT4 E2BTA8 W8BTX9 A0A1B0ARD4 A0A2A3EE21 A0A087ZYZ2 A0A0A1XA21 A0A3L8DVN8 A0A1A9W8E8 A0A034VGS8 A0A151JRU6 A0A195CWK5 F4WFR3 A0A195B1R8 A0A158NYR2 A0A1A9XF06 A0A195FSZ2 B4MYE2 A0A0L0BRB5 A0A1B0ADB8 A0A0M5J4Z9 E2A682 A0A151X2S9 A0A1W4WCG1 B4PAX3 B4KMI5 B3MDL7 B3NQH8 P08841 B4LK54 A0A0K8V413 Q28WU4 A0A336MV72 E9IYR5 A0A194QC42 A0A0J9RJX0 A0A3B0JQE6 A0A0R3NQ23 A0A336MCC6 A0A232EW68 B4J4T7 K7IQN8 A0A1W4W0T8 A0A0B4LGH1 A0A0R1DTQ0 A0A1I8MHC4 B4IHE6 A0A0J9RKH4 A0A212FK03 N6T1W2 A0A1E1VZL2 A0A1I8NYJ0 A0A139WFK6 A0A224XGD3 A0A0V0G361 A0A170XZW9 A0A069DU54 A0A1Y1KNW8 A0A0P4VUI7 R4G588 A0A0K8SM25 R4WCS2 A0A0N0BEN7 A0A067R7H3 V5GSS5
A0A2P8YDV3 A0A182H8I8 A0A1L8E625 A0A1B0C8Q4 A0A182UPL1 A0A182TJQ3 Q17GX8 A0A1S4F2Y4 A0A023EUS0 A0A182QZM6 B0W7F0 A0A182MY89 A0A182MT22 A0A182WUV4 A0A182HYE9 A0A084WAZ5 Q7PPR6 A0A182PJ94 A0A182YF86 A0A182K7P5 A0A182LPX7 A0A182VT65 T1DPP7 A0A182RIT5 A0A182JB51 A0A0L7L7X9 A0A310SK65 A0A1B0G7V4 A0A026WMT3 A0A0L7RDT4 E2BTA8 W8BTX9 A0A1B0ARD4 A0A2A3EE21 A0A087ZYZ2 A0A0A1XA21 A0A3L8DVN8 A0A1A9W8E8 A0A034VGS8 A0A151JRU6 A0A195CWK5 F4WFR3 A0A195B1R8 A0A158NYR2 A0A1A9XF06 A0A195FSZ2 B4MYE2 A0A0L0BRB5 A0A1B0ADB8 A0A0M5J4Z9 E2A682 A0A151X2S9 A0A1W4WCG1 B4PAX3 B4KMI5 B3MDL7 B3NQH8 P08841 B4LK54 A0A0K8V413 Q28WU4 A0A336MV72 E9IYR5 A0A194QC42 A0A0J9RJX0 A0A3B0JQE6 A0A0R3NQ23 A0A336MCC6 A0A232EW68 B4J4T7 K7IQN8 A0A1W4W0T8 A0A0B4LGH1 A0A0R1DTQ0 A0A1I8MHC4 B4IHE6 A0A0J9RKH4 A0A212FK03 N6T1W2 A0A1E1VZL2 A0A1I8NYJ0 A0A139WFK6 A0A224XGD3 A0A0V0G361 A0A170XZW9 A0A069DU54 A0A1Y1KNW8 A0A0P4VUI7 R4G588 A0A0K8SM25 R4WCS2 A0A0N0BEN7 A0A067R7H3 V5GSS5
Pubmed
28756777
12459208
22118469
22651552
26354079
29403074
+ More
26483478 17510324 24945155 24438588 12364791 14747013 17210077 25244985 20966253 26227816 24508170 20798317 24495485 25830018 30249741 25348373 21719571 21347285 17994087 26108605 17550304 3037352 8375576 10731132 12537572 1363225 8485515 15632085 21282665 22936249 28648823 20075255 12537568 12537573 12537574 16110336 17569856 17569867 25315136 23537049 18362917 19820115 26334808 28004739 27129103 23691247 24845553
26483478 17510324 24945155 24438588 12364791 14747013 17210077 25244985 20966253 26227816 24508170 20798317 24495485 25830018 30249741 25348373 21719571 21347285 17994087 26108605 17550304 3037352 8375576 10731132 12537572 1363225 8485515 15632085 21282665 22936249 28648823 20075255 12537568 12537573 12537574 16110336 17569856 17569867 25315136 23537049 18362917 19820115 26334808 28004739 27129103 23691247 24845553
EMBL
ODYU01007605
SOQ50528.1
KZ150040
PZC74569.1
AB072309
BAB86854.1
+ More
AGBW02009721 OWR50037.1 AK401352 BAM17974.1 KQ461072 KPJ09817.1 PYGN01000673 PSN42437.1 JXUM01029849 JXUM01029850 KQ560866 KXJ80565.1 GFDF01000122 JAV13962.1 AJWK01001243 AJWK01001244 AJWK01001245 CH477254 EAT45946.1 GAPW01001384 JAC12214.1 AXCN02002039 DS231853 EDS37865.1 AXCM01000131 APCN01005900 ATLV01022298 KE525331 KFB47389.1 AAAB01008933 EAA09971.5 GAMD01003164 JAA98426.1 JTDY01002354 KOB71598.1 KQ761768 OAD57076.1 CCAG010021503 KK107151 EZA57340.1 KQ414613 KOC68979.1 GL450384 EFN81110.1 GAMC01009814 JAB96741.1 JXJN01002352 JXJN01002353 JXJN01002354 KZ288269 PBC29958.1 GBXI01006779 JAD07513.1 QOIP01000003 RLU24481.1 GAKP01017640 GAKP01017637 JAC41312.1 KQ978566 KYN30108.1 KQ977220 KYN04917.1 GL888122 EGI66956.1 KQ976681 KYM78154.1 ADTU01004211 ADTU01004212 KQ981276 KYN43710.1 CH963894 EDW77131.1 JRES01001473 KNC22620.1 CP012524 ALC41881.1 GL437116 EFN71062.1 KQ982566 KYQ54735.1 CM000158 EDW92513.1 CH933808 EDW10832.1 CH902619 EDV37480.2 CH954179 EDV56981.1 M22335 M22341 X16825 X16826 AE013599 BT015991 X68393 S60740 CH940648 EDW60643.1 GDHF01018766 JAI33548.1 CM000071 EAL26572.1 UFQS01003065 UFQT01003065 SSX15125.1 SSX34504.1 GL767043 EFZ14304.1 KQ459193 KPJ03037.1 CM002911 KMY96308.1 OUUW01000001 SPP73388.1 KRT03176.1 UFQT01000929 SSX28042.1 NNAY01001910 OXU22587.1 CH916367 EDW00633.1 AHN56630.1 KRK00556.1 CH480838 EDW49170.1 KMY96307.1 AGBW02008171 OWR54068.1 APGK01055462 APGK01055463 APGK01055464 APGK01055465 KB741266 ENN71503.1 GDQN01010891 JAT80163.1 KQ971352 KYB26625.1 GFTR01006332 JAW10094.1 GECL01003583 JAP02541.1 GEMB01003848 JAR99406.1 GBGD01001439 JAC87450.1 GEZM01080243 JAV62278.1 GDKW01000385 JAI56210.1 GAHY01000141 JAA77369.1 GBRD01011969 JAG53855.1 AK416954 BAN20169.1 KQ435824 KOX72161.1 KK852686 KDR18432.1 GALX01001232 JAB67234.1
AGBW02009721 OWR50037.1 AK401352 BAM17974.1 KQ461072 KPJ09817.1 PYGN01000673 PSN42437.1 JXUM01029849 JXUM01029850 KQ560866 KXJ80565.1 GFDF01000122 JAV13962.1 AJWK01001243 AJWK01001244 AJWK01001245 CH477254 EAT45946.1 GAPW01001384 JAC12214.1 AXCN02002039 DS231853 EDS37865.1 AXCM01000131 APCN01005900 ATLV01022298 KE525331 KFB47389.1 AAAB01008933 EAA09971.5 GAMD01003164 JAA98426.1 JTDY01002354 KOB71598.1 KQ761768 OAD57076.1 CCAG010021503 KK107151 EZA57340.1 KQ414613 KOC68979.1 GL450384 EFN81110.1 GAMC01009814 JAB96741.1 JXJN01002352 JXJN01002353 JXJN01002354 KZ288269 PBC29958.1 GBXI01006779 JAD07513.1 QOIP01000003 RLU24481.1 GAKP01017640 GAKP01017637 JAC41312.1 KQ978566 KYN30108.1 KQ977220 KYN04917.1 GL888122 EGI66956.1 KQ976681 KYM78154.1 ADTU01004211 ADTU01004212 KQ981276 KYN43710.1 CH963894 EDW77131.1 JRES01001473 KNC22620.1 CP012524 ALC41881.1 GL437116 EFN71062.1 KQ982566 KYQ54735.1 CM000158 EDW92513.1 CH933808 EDW10832.1 CH902619 EDV37480.2 CH954179 EDV56981.1 M22335 M22341 X16825 X16826 AE013599 BT015991 X68393 S60740 CH940648 EDW60643.1 GDHF01018766 JAI33548.1 CM000071 EAL26572.1 UFQS01003065 UFQT01003065 SSX15125.1 SSX34504.1 GL767043 EFZ14304.1 KQ459193 KPJ03037.1 CM002911 KMY96308.1 OUUW01000001 SPP73388.1 KRT03176.1 UFQT01000929 SSX28042.1 NNAY01001910 OXU22587.1 CH916367 EDW00633.1 AHN56630.1 KRK00556.1 CH480838 EDW49170.1 KMY96307.1 AGBW02008171 OWR54068.1 APGK01055462 APGK01055463 APGK01055464 APGK01055465 KB741266 ENN71503.1 GDQN01010891 JAT80163.1 KQ971352 KYB26625.1 GFTR01006332 JAW10094.1 GECL01003583 JAP02541.1 GEMB01003848 JAR99406.1 GBGD01001439 JAC87450.1 GEZM01080243 JAV62278.1 GDKW01000385 JAI56210.1 GAHY01000141 JAA77369.1 GBRD01011969 JAG53855.1 AK416954 BAN20169.1 KQ435824 KOX72161.1 KK852686 KDR18432.1 GALX01001232 JAB67234.1
Proteomes
UP000007151
UP000053240
UP000245037
UP000069940
UP000249989
UP000092461
+ More
UP000075903 UP000075902 UP000008820 UP000075886 UP000002320 UP000075884 UP000075883 UP000076407 UP000075840 UP000030765 UP000007062 UP000075885 UP000076408 UP000075881 UP000075882 UP000075920 UP000075900 UP000075880 UP000037510 UP000092444 UP000053097 UP000053825 UP000008237 UP000092460 UP000242457 UP000005203 UP000279307 UP000091820 UP000078492 UP000078542 UP000007755 UP000078540 UP000005205 UP000092443 UP000078541 UP000007798 UP000037069 UP000092445 UP000092553 UP000000311 UP000075809 UP000192221 UP000002282 UP000009192 UP000007801 UP000008711 UP000000803 UP000008792 UP000001819 UP000053268 UP000268350 UP000215335 UP000001070 UP000002358 UP000095301 UP000001292 UP000019118 UP000095300 UP000007266 UP000053105 UP000027135
UP000075903 UP000075902 UP000008820 UP000075886 UP000002320 UP000075884 UP000075883 UP000076407 UP000075840 UP000030765 UP000007062 UP000075885 UP000076408 UP000075881 UP000075882 UP000075920 UP000075900 UP000075880 UP000037510 UP000092444 UP000053097 UP000053825 UP000008237 UP000092460 UP000242457 UP000005203 UP000279307 UP000091820 UP000078492 UP000078542 UP000007755 UP000078540 UP000005205 UP000092443 UP000078541 UP000007798 UP000037069 UP000092445 UP000092553 UP000000311 UP000075809 UP000192221 UP000002282 UP000009192 UP000007801 UP000008711 UP000000803 UP000008792 UP000001819 UP000053268 UP000268350 UP000215335 UP000001070 UP000002358 UP000095301 UP000001292 UP000019118 UP000095300 UP000007266 UP000053105 UP000027135
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2H1WBT4
A0A2W1BHJ4
Q8T8B1
A0A212F8H2
I4DJ84
A0A0N1INH4
+ More
A0A2P8YDV3 A0A182H8I8 A0A1L8E625 A0A1B0C8Q4 A0A182UPL1 A0A182TJQ3 Q17GX8 A0A1S4F2Y4 A0A023EUS0 A0A182QZM6 B0W7F0 A0A182MY89 A0A182MT22 A0A182WUV4 A0A182HYE9 A0A084WAZ5 Q7PPR6 A0A182PJ94 A0A182YF86 A0A182K7P5 A0A182LPX7 A0A182VT65 T1DPP7 A0A182RIT5 A0A182JB51 A0A0L7L7X9 A0A310SK65 A0A1B0G7V4 A0A026WMT3 A0A0L7RDT4 E2BTA8 W8BTX9 A0A1B0ARD4 A0A2A3EE21 A0A087ZYZ2 A0A0A1XA21 A0A3L8DVN8 A0A1A9W8E8 A0A034VGS8 A0A151JRU6 A0A195CWK5 F4WFR3 A0A195B1R8 A0A158NYR2 A0A1A9XF06 A0A195FSZ2 B4MYE2 A0A0L0BRB5 A0A1B0ADB8 A0A0M5J4Z9 E2A682 A0A151X2S9 A0A1W4WCG1 B4PAX3 B4KMI5 B3MDL7 B3NQH8 P08841 B4LK54 A0A0K8V413 Q28WU4 A0A336MV72 E9IYR5 A0A194QC42 A0A0J9RJX0 A0A3B0JQE6 A0A0R3NQ23 A0A336MCC6 A0A232EW68 B4J4T7 K7IQN8 A0A1W4W0T8 A0A0B4LGH1 A0A0R1DTQ0 A0A1I8MHC4 B4IHE6 A0A0J9RKH4 A0A212FK03 N6T1W2 A0A1E1VZL2 A0A1I8NYJ0 A0A139WFK6 A0A224XGD3 A0A0V0G361 A0A170XZW9 A0A069DU54 A0A1Y1KNW8 A0A0P4VUI7 R4G588 A0A0K8SM25 R4WCS2 A0A0N0BEN7 A0A067R7H3 V5GSS5
A0A2P8YDV3 A0A182H8I8 A0A1L8E625 A0A1B0C8Q4 A0A182UPL1 A0A182TJQ3 Q17GX8 A0A1S4F2Y4 A0A023EUS0 A0A182QZM6 B0W7F0 A0A182MY89 A0A182MT22 A0A182WUV4 A0A182HYE9 A0A084WAZ5 Q7PPR6 A0A182PJ94 A0A182YF86 A0A182K7P5 A0A182LPX7 A0A182VT65 T1DPP7 A0A182RIT5 A0A182JB51 A0A0L7L7X9 A0A310SK65 A0A1B0G7V4 A0A026WMT3 A0A0L7RDT4 E2BTA8 W8BTX9 A0A1B0ARD4 A0A2A3EE21 A0A087ZYZ2 A0A0A1XA21 A0A3L8DVN8 A0A1A9W8E8 A0A034VGS8 A0A151JRU6 A0A195CWK5 F4WFR3 A0A195B1R8 A0A158NYR2 A0A1A9XF06 A0A195FSZ2 B4MYE2 A0A0L0BRB5 A0A1B0ADB8 A0A0M5J4Z9 E2A682 A0A151X2S9 A0A1W4WCG1 B4PAX3 B4KMI5 B3MDL7 B3NQH8 P08841 B4LK54 A0A0K8V413 Q28WU4 A0A336MV72 E9IYR5 A0A194QC42 A0A0J9RJX0 A0A3B0JQE6 A0A0R3NQ23 A0A336MCC6 A0A232EW68 B4J4T7 K7IQN8 A0A1W4W0T8 A0A0B4LGH1 A0A0R1DTQ0 A0A1I8MHC4 B4IHE6 A0A0J9RKH4 A0A212FK03 N6T1W2 A0A1E1VZL2 A0A1I8NYJ0 A0A139WFK6 A0A224XGD3 A0A0V0G361 A0A170XZW9 A0A069DU54 A0A1Y1KNW8 A0A0P4VUI7 R4G588 A0A0K8SM25 R4WCS2 A0A0N0BEN7 A0A067R7H3 V5GSS5
PDB
5KMG
E-value=0,
Score=1965
Ontologies
GO
PANTHER
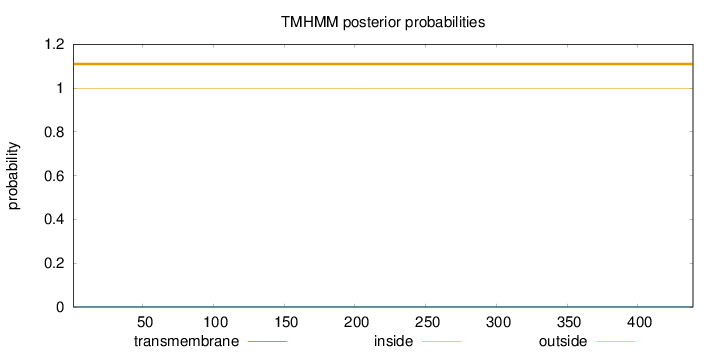
Topology
Subcellular location
Length:
439
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00514999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00180
outside
1 - 439
Population Genetic Test Statistics
Pi
25.512128
Theta
28.082212
Tajima's D
-0.46058
CLR
0.054919
CSRT
0.250287485625719
Interpretation
Uncertain
