Gene
KWMTBOMO12280
Pre Gene Modal
BGIBMGA009134
Annotation
PREDICTED:_activin_receptor_type-1_[Amyelois_transitella]
Full name
Receptor protein serine/threonine kinase
Location in the cell
Nuclear Reliability : 2.021
Sequence
CDS
ATGGCGGACTCTGTCATATTTGGATATCTGTTCATATTTCTGTTGATAAAGCCTAACTATGCAAGTCTCCCCAATTTGGATGCATTGGTCCAAGATGGACAGGGCAGGGTCCTACAGCAATTGGAAGATCCGATCACACTCGAAAAGATGATGGTACACCCCCCAAAATACAAATGTCACCTCTGCGAGCCCCCTGACTGCGAATCTGGTGTCCAGAACATATGCGTCGACGCCCTATATTGCTTCAAGTCCTCGGTGAGGGAGTCCAATGGAGAGCTGCAGCACTCAAGGGGTTGCTCCGATAAGAACGATCACTACCAGTTCATATGTCGCTCAAGTACGAGGCAAGGGGTCCACAAGCGTCACGCCGGTCAGCTGAACGTGACGTGCTGCACGGGGGACATGTGCAACTCGGGCGAGTTCCCGGCGCTGTGGGAGGCGGCGCGGGCGGGGCGCGCGGCCTGGGAGCCGTGGGCGGGCGGCGCCGCGGCCGTGCTGCTGCTGGCGGCGCTGCTGACGGGCCTGCTGCTGCTGCTGCAACGAGCCCACAGGCTGCGCGTGTCGCGGACTGCCCTGGACAAACTCCGCTCCGAGGCGAAGTACTTCTCATTTAACGTCTACAACCAGCACATGAACAACGCTTATTACGAAGGTGCGGAGGGCTCACAGCCGGGCTGCGTGGCGGTGGCGGCCGGGGACTCCACGCTGCGCGAGTACCTGGAGGGCTCCCTGAGCTCGGGCTCCGGCTCGGGCCTGCCGCTGATGGTGCAGCGCACGCTCGCCAAGCAGGTCACGCTGTACGACTGCGTCGGCAAGGGCCGCTACGGCGAGGTGTGGCGCGGCTCGTGGTACGGGGACAGCGTGGCGGTCAAGATCTTCTTCTCGCGAGACGAGGCCAGCTGGAAGCGCGAGACCGAGGTGTACAGCACCGTGCTGCTGCGCCACCAGAACATCCTGGCCTACGTGGGCAGCGACATGACCAGCCGCGGCAGCTGCACGCAGCTGTGGCTGATCACGCAGTACCACCCGCTGGGCTCGCTGCACGAGCACCTGCAGCGCGGCACGCTCTCCCGCCAGCAGCTCGCCGCGCTCAGCCTGTCCGCCGCCAGCGGCCTGCTGCACCTGCACACGCAGATACACGGCACCCAGGGTAAACCTGCGATAGCACACCGGGACATCAAGTCCAAGAACATCCTGGTCAAGGTGAATGGGGAGTGTTGCATCGGAGACCTCGGGCTCGCCCTCACCGCGCAGCACATCGAGCAGGGCCAGCAGCTGCACAACCCTCGCCAGGGGACCAAGAGGTATATGAGCCCCGAGATGCTGGACCACACCATGCAGACGGACTGCCTGGACGCGTACCTGCGCTGCGACGTGTACGCGCTGGGGCTGGTGCTGTGGGAGCTGTGCCGCCGCGTGCCCCCCGCGCGCCCGCCCGCGCCCCCCTACCACCACCGCGCGCCGCCCGACCCCTCCTACGAGGACATGCGCAAGATCGTCTGCGTGGACCGGGACCGACCCGAGCCGCTGGACTACCACGATCATCCGACAATGGTCTCAATGTGGAAACTGATCCAAGAGTGCTGGCACCACAACCCGTCGGCGCGCCTCTCCGCGCTGCGCGTCAGGAAGACCTTACTGAAGATTGCAGCAGCGGACGAGTCCATACACCTCAACGAGGACAACGAGGTGTCCCTGTAG
Protein
MADSVIFGYLFIFLLIKPNYASLPNLDALVQDGQGRVLQQLEDPITLEKMMVHPPKYKCHLCEPPDCESGVQNICVDALYCFKSSVRESNGELQHSRGCSDKNDHYQFICRSSTRQGVHKRHAGQLNVTCCTGDMCNSGEFPALWEAARAGRAAWEPWAGGAAAVLLLAALLTGLLLLLQRAHRLRVSRTALDKLRSEAKYFSFNVYNQHMNNAYYEGAEGSQPGCVAVAAGDSTLREYLEGSLSSGSGSGLPLMVQRTLAKQVTLYDCVGKGRYGEVWRGSWYGDSVAVKIFFSRDEASWKRETEVYSTVLLRHQNILAYVGSDMTSRGSCTQLWLITQYHPLGSLHEHLQRGTLSRQQLAALSLSAASGLLHLHTQIHGTQGKPAIAHRDIKSKNILVKVNGECCIGDLGLALTAQHIEQGQQLHNPRQGTKRYMSPEMLDHTMQTDCLDAYLRCDVYALGLVLWELCRRVPPARPPAPPYHHRAPPDPSYEDMRKIVCVDRDRPEPLDYHDHPTMVSMWKLIQECWHHNPSARLSALRVRKTLLKIAAADESIHLNEDNEVSL
Summary
Catalytic Activity
[receptor-protein]-L-serine + ATP = [receptor-protein]-O-phospho-L-serine + ADP + H(+)
[receptor-protein]-L-threonine + ATP = [receptor-protein]-O-phospho-L-threonine + ADP + H(+)
[receptor-protein]-L-threonine + ATP = [receptor-protein]-O-phospho-L-threonine + ADP + H(+)
Similarity
Belongs to the protein kinase superfamily. TKL Ser/Thr protein kinase family. TGFB receptor subfamily.
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily.
Feature
chain Receptor protein serine/threonine kinase
Uniprot
H9JHY6
A0A2W1BPA6
A0A2H1WBV9
A0A0N0PB92
A0A194QDJ8
A0A212F8J5
+ More
E0W0F7 A0A1Y1KSQ7 C9QP22 Q7JQ36 H5V885 Q7JPM7 Q7JPN8 A0A0J9R7Q9 B4HR79 A0A0T6AVB4 B3N9E3 Q7JQ39 B4QES9 A0A0J9R7H5 A0A1Q3FX50 A0A3B0JM54 A0A1Q3FX39 Q95U21 B4GDK4 Q292G4 D5SHT1 A0A0B4K7J3 B4P239 B4NS66 A0A084VF32 A0A0J9R8S8 A0A182V796 A0A0R3NL31 B4MYH3 A0A1W4WBG3 A0A1B6BYG4 A0A182GZD7 A0A1W4VZ96 Q7QJY6 Q16MX6 B4J4Y1 B3MG92 A0A1W4VZ93 A0A0M4E498 A0A0N8P0G2 B4LMK2 A0A1S4FVA0 A0A023EU41 A0A182NH98 A0A182QAU5 A0A182JN48 A0A182JT42 B4KSY4 A0A0Q9WFH1 A0A1I8Q056 A0A0Q9XIJ8 A0A1I8Q057 A0A0P5KBW8 E9HSW3 A0A0P6DRD4 A0A0P6E301 A0A182LGT8 A0A0F7ZBD9 A0A3Q3DJJ0
E0W0F7 A0A1Y1KSQ7 C9QP22 Q7JQ36 H5V885 Q7JPM7 Q7JPN8 A0A0J9R7Q9 B4HR79 A0A0T6AVB4 B3N9E3 Q7JQ39 B4QES9 A0A0J9R7H5 A0A1Q3FX50 A0A3B0JM54 A0A1Q3FX39 Q95U21 B4GDK4 Q292G4 D5SHT1 A0A0B4K7J3 B4P239 B4NS66 A0A084VF32 A0A0J9R8S8 A0A182V796 A0A0R3NL31 B4MYH3 A0A1W4WBG3 A0A1B6BYG4 A0A182GZD7 A0A1W4VZ96 Q7QJY6 Q16MX6 B4J4Y1 B3MG92 A0A1W4VZ93 A0A0M4E498 A0A0N8P0G2 B4LMK2 A0A1S4FVA0 A0A023EU41 A0A182NH98 A0A182QAU5 A0A182JN48 A0A182JT42 B4KSY4 A0A0Q9WFH1 A0A1I8Q056 A0A0Q9XIJ8 A0A1I8Q057 A0A0P5KBW8 E9HSW3 A0A0P6DRD4 A0A0P6E301 A0A182LGT8 A0A0F7ZBD9 A0A3Q3DJJ0
EC Number
2.7.11.30
Pubmed
EMBL
BABH01035348
KZ150040
PZC74570.1
ODYU01007605
SOQ50527.1
KQ461072
+ More
KPJ09816.1 KQ459193 KPJ03040.1 AGBW02009721 OWR50038.1 DS235858 EEB19113.1 GEZM01074879 JAV64423.1 BT099904 ACX32975.1 L33790 AE013599 AAA61946.1 AAF59189.1 BT133186 AFA28427.1 U11441 AAA53242.1 U05209 AAA18208.1 CM002911 KMY92098.1 CH480816 EDW46822.1 LJIG01022730 KRT79009.1 CH954177 EDV59630.1 L33476 AAA28878.1 CM000362 EDX06075.1 KMY92097.1 GFDL01002886 JAV32159.1 OUUW01000001 SPP74356.1 GFDL01003023 JAV32022.1 AY058363 AAL13592.1 AAM68863.1 CH479181 EDW31661.1 CM000071 EAL24898.1 BT124882 ADG46050.1 AFH07948.1 CM000157 EDW89240.1 CH981919 EDX15444.1 ATLV01012360 KE524785 KFB36576.1 KMY92099.1 KRT01656.1 CH963894 EDW77162.1 GEDC01030985 JAS06313.1 JXUM01099363 KQ564486 KXJ72211.1 AAAB01008807 EAA04704.4 CH477846 EAT35698.1 CH916367 EDW01687.1 CH902619 EDV37795.1 CP012524 ALC40816.1 KPU77036.1 CH940648 EDW59989.2 GAPW01000705 JAC12893.1 AXCN02000894 CH933808 EDW08481.2 KRF79184.1 KRG04145.1 GDIQ01212295 JAK39430.1 GL732759 EFX65169.1 GDIQ01073497 JAN21240.1 GDIQ01073498 JAN21239.1 GBEX01000080 JAI14480.1
KPJ09816.1 KQ459193 KPJ03040.1 AGBW02009721 OWR50038.1 DS235858 EEB19113.1 GEZM01074879 JAV64423.1 BT099904 ACX32975.1 L33790 AE013599 AAA61946.1 AAF59189.1 BT133186 AFA28427.1 U11441 AAA53242.1 U05209 AAA18208.1 CM002911 KMY92098.1 CH480816 EDW46822.1 LJIG01022730 KRT79009.1 CH954177 EDV59630.1 L33476 AAA28878.1 CM000362 EDX06075.1 KMY92097.1 GFDL01002886 JAV32159.1 OUUW01000001 SPP74356.1 GFDL01003023 JAV32022.1 AY058363 AAL13592.1 AAM68863.1 CH479181 EDW31661.1 CM000071 EAL24898.1 BT124882 ADG46050.1 AFH07948.1 CM000157 EDW89240.1 CH981919 EDX15444.1 ATLV01012360 KE524785 KFB36576.1 KMY92099.1 KRT01656.1 CH963894 EDW77162.1 GEDC01030985 JAS06313.1 JXUM01099363 KQ564486 KXJ72211.1 AAAB01008807 EAA04704.4 CH477846 EAT35698.1 CH916367 EDW01687.1 CH902619 EDV37795.1 CP012524 ALC40816.1 KPU77036.1 CH940648 EDW59989.2 GAPW01000705 JAC12893.1 AXCN02000894 CH933808 EDW08481.2 KRF79184.1 KRG04145.1 GDIQ01212295 JAK39430.1 GL732759 EFX65169.1 GDIQ01073497 JAN21240.1 GDIQ01073498 JAN21239.1 GBEX01000080 JAI14480.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000009046
UP000000803
+ More
UP000001292 UP000008711 UP000000304 UP000268350 UP000008744 UP000001819 UP000002282 UP000030765 UP000075903 UP000007798 UP000192221 UP000069940 UP000249989 UP000007062 UP000008820 UP000001070 UP000007801 UP000092553 UP000008792 UP000075884 UP000075886 UP000075880 UP000075881 UP000009192 UP000095300 UP000000305 UP000075882 UP000264820
UP000001292 UP000008711 UP000000304 UP000268350 UP000008744 UP000001819 UP000002282 UP000030765 UP000075903 UP000007798 UP000192221 UP000069940 UP000249989 UP000007062 UP000008820 UP000001070 UP000007801 UP000092553 UP000008792 UP000075884 UP000075886 UP000075880 UP000075881 UP000009192 UP000095300 UP000000305 UP000075882 UP000264820
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9JHY6
A0A2W1BPA6
A0A2H1WBV9
A0A0N0PB92
A0A194QDJ8
A0A212F8J5
+ More
E0W0F7 A0A1Y1KSQ7 C9QP22 Q7JQ36 H5V885 Q7JPM7 Q7JPN8 A0A0J9R7Q9 B4HR79 A0A0T6AVB4 B3N9E3 Q7JQ39 B4QES9 A0A0J9R7H5 A0A1Q3FX50 A0A3B0JM54 A0A1Q3FX39 Q95U21 B4GDK4 Q292G4 D5SHT1 A0A0B4K7J3 B4P239 B4NS66 A0A084VF32 A0A0J9R8S8 A0A182V796 A0A0R3NL31 B4MYH3 A0A1W4WBG3 A0A1B6BYG4 A0A182GZD7 A0A1W4VZ96 Q7QJY6 Q16MX6 B4J4Y1 B3MG92 A0A1W4VZ93 A0A0M4E498 A0A0N8P0G2 B4LMK2 A0A1S4FVA0 A0A023EU41 A0A182NH98 A0A182QAU5 A0A182JN48 A0A182JT42 B4KSY4 A0A0Q9WFH1 A0A1I8Q056 A0A0Q9XIJ8 A0A1I8Q057 A0A0P5KBW8 E9HSW3 A0A0P6DRD4 A0A0P6E301 A0A182LGT8 A0A0F7ZBD9 A0A3Q3DJJ0
E0W0F7 A0A1Y1KSQ7 C9QP22 Q7JQ36 H5V885 Q7JPM7 Q7JPN8 A0A0J9R7Q9 B4HR79 A0A0T6AVB4 B3N9E3 Q7JQ39 B4QES9 A0A0J9R7H5 A0A1Q3FX50 A0A3B0JM54 A0A1Q3FX39 Q95U21 B4GDK4 Q292G4 D5SHT1 A0A0B4K7J3 B4P239 B4NS66 A0A084VF32 A0A0J9R8S8 A0A182V796 A0A0R3NL31 B4MYH3 A0A1W4WBG3 A0A1B6BYG4 A0A182GZD7 A0A1W4VZ96 Q7QJY6 Q16MX6 B4J4Y1 B3MG92 A0A1W4VZ93 A0A0M4E498 A0A0N8P0G2 B4LMK2 A0A1S4FVA0 A0A023EU41 A0A182NH98 A0A182QAU5 A0A182JN48 A0A182JT42 B4KSY4 A0A0Q9WFH1 A0A1I8Q056 A0A0Q9XIJ8 A0A1I8Q057 A0A0P5KBW8 E9HSW3 A0A0P6DRD4 A0A0P6E301 A0A182LGT8 A0A0F7ZBD9 A0A3Q3DJJ0
PDB
4C02
E-value=7.2129e-97,
Score=905
Ontologies
GO
GO:0005524
GO:0016021
GO:0004675
GO:0007300
GO:0007179
GO:0007448
GO:0001745
GO:0035222
GO:0030512
GO:0006468
GO:0004672
GO:0045887
GO:0007476
GO:0005025
GO:0030718
GO:0045824
GO:0042078
GO:0050829
GO:0030707
GO:0030514
GO:0008358
GO:0070724
GO:0009953
GO:0036122
GO:0098821
GO:0043235
GO:0046872
GO:0007389
GO:0048179
GO:0005886
GO:0030509
GO:0046332
GO:0005887
GO:0019838
GO:0048185
GO:0005024
GO:0032924
GO:0004674
GO:0016020
GO:0019538
GO:0030145
GO:0000049
GO:0005975
GO:0004553
PANTHER
Topology
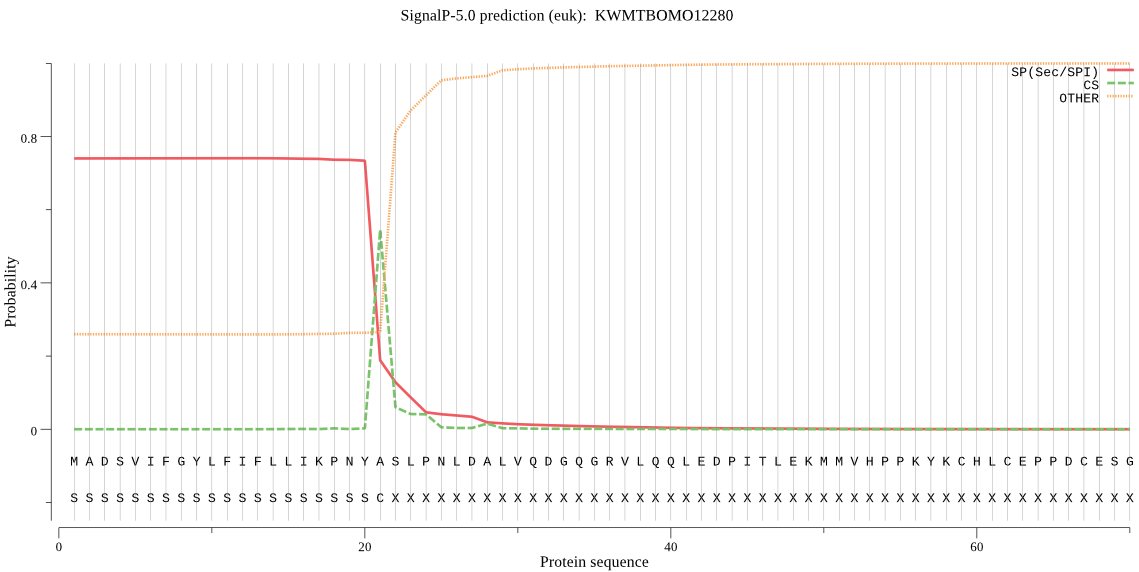
SignalP
Position: 1 - 21,
Likelihood: 0.741449
Length:
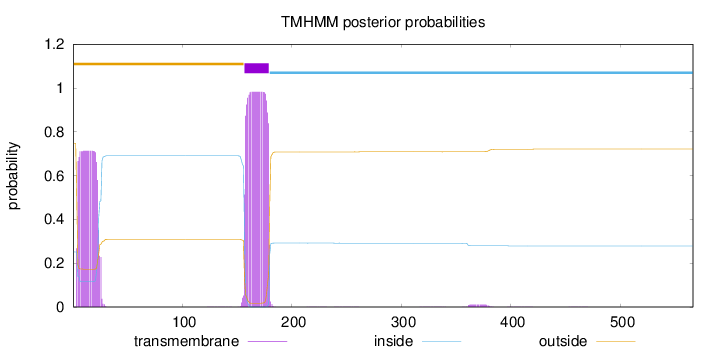
566
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
36.7722500000001
Exp number, first 60 AAs:
14.41585
Total prob of N-in:
0.25219
POSSIBLE N-term signal
sequence
outside
1 - 156
TMhelix
157 - 179
inside
180 - 566
Population Genetic Test Statistics
Pi
23.951466
Theta
26.682717
Tajima's D
-0.401143
CLR
1.297532
CSRT
0.268286585670716
Interpretation
Uncertain
