Gene
KWMTBOMO12266
Pre Gene Modal
BGIBMGA009136
Annotation
PREDICTED:_lipase_member_H-B-like_[Bombyx_mori]
Full name
Lipase member H-B
+ More
Pancreatic lipase-related protein 2
Triacylglycerol lipase
Lipase member H-A
Pancreatic lipase-related protein 2
Triacylglycerol lipase
Lipase member H-A
Alternative Name
GPL
Galactolipase
Pancreatic lipase
Galactolipase
Pancreatic lipase
Location in the cell
PlasmaMembrane Reliability : 1.955
Sequence
CDS
ATGGATCCGAGGAGTGAGACTGGATATCCTACGGGAATGATGTCTGTTTGTCCCGGCTCCACGAAGCCTGCAATCATACCGAAGAGCCAGCTGAAGTACCTGTCCTTCGTGGTGCAGGGGAACGGGAGAAGCCGGCGCAAGTACTCATATTGGAACGCTAGGAATATAGCCAAGGATCCGAGAATCAATTTTAAAAGGAAAACTCTCCTCCTGGCCATAGGGTATCTCGACAGCCCCAGTTTCCCCATTTCAATGATGTTCGCGAACGAGTACGAAGCTCTGGGGTACAATGTCATAATTATTGACAACCAGAGGTTTTCGACTGTGCATTACCATTTAGCAAGCCGCCTCATGAGACCAGTGGGTAAGCTTGTAGCCGAAGTCCTAGTCCAGCTAACTGACTACGGCCTAGACCCTTCCAGGCTAGAATTATTAGGGTTTAGTCTGGGCGCCCAAACGGTCAGCTACATAGCCAAAAACTTCCAGACCATAACAGGAAGGAACGTATCGTCTATCGTCGCTCTAGAGCCTTCCGGCCCGTGTTTTAGGACATTAGTTTCGGACGACAGACTCAACCCGTCAGACGCTGGCTTCGTGCAGGTCGTGCACACGAACATAGACGGGTACGGTATGGCCAACAGAATGGGCCACGTCGACTTCTACGTCAACGGCGGGGAGTACCAACCCTCAGACATCAACCTTTACCCTTGTACGACAACTTGCAGCCATTTCAGGGTGCTCGCGCTTTGGGTTGCCGCCTTAAAGAATCCCGGTAAATTTATAGGTATGAAGTGTGATAGTGTGCAGCAAGCGAGGGACGCTCAGTGCTATAACAACAAGCCTCTCGTCACTAATACAATGGGTTTGAGTCCTGACAAGAGTAAGCTGGGCATATTTTATTTATCGACTAGTAAGGGGTATCCTTACTTCTTGGGGGCAAAAGGACTGATAGAGGCCAATGTTTACTGGCGGACGATTACTGAAATCAATAATGGTGACGATATTTTAGTGTACACTTAA
Protein
MDPRSETGYPTGMMSVCPGSTKPAIIPKSQLKYLSFVVQGNGRSRRKYSYWNARNIAKDPRINFKRKTLLLAIGYLDSPSFPISMMFANEYEALGYNVIIIDNQRFSTVHYHLASRLMRPVGKLVAEVLVQLTDYGLDPSRLELLGFSLGAQTVSYIAKNFQTITGRNVSSIVALEPSGPCFRTLVSDDRLNPSDAGFVQVVHTNIDGYGMANRMGHVDFYVNGGEYQPSDINLYPCTTTCSHFRVLALWVAALKNPGKFIGMKCDSVQQARDAQCYNNKPLVTNTMGLSPDKSKLGIFYLSTSKGYPYFLGAKGLIEANVYWRTITEINNGDDILVYT
Summary
Description
Hydrolyzes specifically phosphatidic acid (PA) to produce lysophosphatidic acid (LPA).
Lipase with broad substrate specificity. Can hydrolyze both phospholipids and galactolipids. Acts preferentially on monoglycerides, phospholipids and galactolipids. Contributes to milk fat hydrolysis (By similarity).
Lipase with broad substrate specificity. Can hydrolyze both phospholipids and galactolipids. Acts preferentially on monoglycerides, phospholipids and galactolipids. Contributes to milk fat hydrolysis (By similarity).
Catalytic Activity
a triacylglycerol + H2O = a diacylglycerol + a fatty acid + H(+)
a 1,2-diacyl-3-O-(beta-D-galactosyl)-sn-glycerol + 2 H2O = 3-beta-D-galactosyl-sn-glycerol + 2 a fatty acid + 2 H(+)
a 1,2-diacyl-3-O-(beta-D-galactosyl)-sn-glycerol + 2 H2O = 3-beta-D-galactosyl-sn-glycerol + 2 a fatty acid + 2 H(+)
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Keywords
Disulfide bond
Glycoprotein
Hydrolase
Lipid degradation
Lipid metabolism
Membrane
Secreted
Signal
3D-structure
Calcium
Complete proteome
Direct protein sequencing
Metal-binding
Reference proteome
Feature
chain Lipase member H-B
Uniprot
H9JHY8
A0A2H1WL24
A0A2W1BNT7
A0A194QBX2
A0A194QUH6
A0A212FPW6
+ More
A0A0L7L1G4 A0A194Q1D6 A0A0N1II42 A0A2W1B572 A0A212FBR8 H9JQ12 A0A2H1WK50 H9JQQ8 A0A2A4JBH9 A0A191XQW7 A0A3S2LBT4 A0A2H1VJ13 A0A0L7LHY6 A0A194QBZ0 A0A0N1IGL3 A0A3S2PK48 A0A2A4JXD4 A0A2W1BL36 A0A194PEP3 A0A3S2P6E3 A0A194QZ66 A0A2H1VGU5 A0A212EY15 A0A212EJL3 A0A2W1BWG2 A0A2A4JGD9 A0A2H1X3Q9 A0A3S2NZZ7 H9JWY8 A0A3S2L3L9 A0A2W1BV63 A0A2H1W795 A0A0L7KP71 I6TFN4 A0A2W1BXY3 A0A220K8L9 A0A2H1W7F1 A0A0L7L8R4 A0A194PCA8 A0A2H1WNR8 A0A194QNS4 A0A2H1WL84 A0A2A4IT83 A0A2A4J1M9 A0A210QK37 A0A1B6MCF6 A0A212FKA5 A0A1B6L5N2 A0A194QC74 Q641F6 A0A210PSA1 A0A1L8H505 P81139 A0A2H1W9L5 H0V008 T1H7M9 A0A224XHR8 A0A1B0G9P9 A0A2B4RJ44 A0A3B3ZZW1 Q6PA23 A0A2W1BTP2 A0A1B0AP02 A0A1L8HCT6 A0A3Q3WX63 A0A2W1B7Q2 D6W6G8 A0A0L7LP71 A0A1W4WIH6 K7GJB0
A0A0L7L1G4 A0A194Q1D6 A0A0N1II42 A0A2W1B572 A0A212FBR8 H9JQ12 A0A2H1WK50 H9JQQ8 A0A2A4JBH9 A0A191XQW7 A0A3S2LBT4 A0A2H1VJ13 A0A0L7LHY6 A0A194QBZ0 A0A0N1IGL3 A0A3S2PK48 A0A2A4JXD4 A0A2W1BL36 A0A194PEP3 A0A3S2P6E3 A0A194QZ66 A0A2H1VGU5 A0A212EY15 A0A212EJL3 A0A2W1BWG2 A0A2A4JGD9 A0A2H1X3Q9 A0A3S2NZZ7 H9JWY8 A0A3S2L3L9 A0A2W1BV63 A0A2H1W795 A0A0L7KP71 I6TFN4 A0A2W1BXY3 A0A220K8L9 A0A2H1W7F1 A0A0L7L8R4 A0A194PCA8 A0A2H1WNR8 A0A194QNS4 A0A2H1WL84 A0A2A4IT83 A0A2A4J1M9 A0A210QK37 A0A1B6MCF6 A0A212FKA5 A0A1B6L5N2 A0A194QC74 Q641F6 A0A210PSA1 A0A1L8H505 P81139 A0A2H1W9L5 H0V008 T1H7M9 A0A224XHR8 A0A1B0G9P9 A0A2B4RJ44 A0A3B3ZZW1 Q6PA23 A0A2W1BTP2 A0A1B0AP02 A0A1L8HCT6 A0A3Q3WX63 A0A2W1B7Q2 D6W6G8 A0A0L7LP71 A0A1W4WIH6 K7GJB0
EC Number
3.1.1.-
3.1.1.3
3.1.1.3
Pubmed
EMBL
BABH01035306
ODYU01009344
SOQ53707.1
KZ149947
PZC76719.1
KQ459193
+ More
KPJ03053.1 KQ461154 KPJ08630.1 AGBW02000859 OWR55743.1 JTDY01003560 KOB69343.1 KQ459590 KPI97210.1 KQ460391 KPJ15413.1 KZ150534 PZC70591.1 AGBW02009288 OWR51181.1 BABH01012334 ODYU01009136 SOQ53346.1 NWSH01002209 PCG68904.1 KU360126 ANJ42864.1 RSAL01000052 RVE50204.1 ODYU01002823 SOQ40786.1 JTDY01001036 KOB75067.1 KPJ03068.1 KQ460781 KPJ12226.1 RSAL01000010 RVE53725.1 NWSH01000400 PCG76675.1 KZ149983 PZC75749.1 KQ459606 KPI91164.1 RSAL01000291 RVE42897.1 KQ460947 KPJ10569.1 ODYU01002500 SOQ40069.1 AGBW02011630 OWR46389.1 AGBW02014435 OWR41693.1 KZ149926 PZC77547.1 NWSH01001670 PCG70462.1 ODYU01013233 SOQ59929.1 RSAL01000012 RVE53468.1 BABH01041781 RSAL01000184 RVE44850.1 PZC77545.1 ODYU01006796 SOQ48975.1 JTDY01007594 KOB65092.1 JQ814807 AFM73623.1 PZC77546.1 MF319718 ASJ26432.1 SOQ48977.1 JTDY01002277 KOB71750.1 KPI90911.1 ODYU01009953 SOQ54715.1 KQ461196 KPJ06625.1 ODYU01009427 SOQ53845.1 NWSH01007523 PCG62931.1 NWSH01003717 PCG65891.1 NEDP02003256 OWF49099.1 GEBQ01006364 JAT33613.1 AGBW02008082 OWR54177.1 GEBQ01020948 JAT19029.1 KPJ03024.1 BC082381 NEDP02005531 OWF39326.1 CM004469 OCT91179.1 ODYU01007212 SOQ49795.1 AAKN02003095 ACPB03000883 GFTR01007078 JAW09348.1 CCAG010003515 LSMT01000515 PFX16839.1 BC060482 KZ149921 PZC77721.1 JXJN01001096 CM004468 OCT93917.1 KZ150304 PZC71518.1 KQ971307 EFA11062.1 JTDY01000429 KOB77220.1 AGCU01091550 AGCU01091551 AGCU01091552
KPJ03053.1 KQ461154 KPJ08630.1 AGBW02000859 OWR55743.1 JTDY01003560 KOB69343.1 KQ459590 KPI97210.1 KQ460391 KPJ15413.1 KZ150534 PZC70591.1 AGBW02009288 OWR51181.1 BABH01012334 ODYU01009136 SOQ53346.1 NWSH01002209 PCG68904.1 KU360126 ANJ42864.1 RSAL01000052 RVE50204.1 ODYU01002823 SOQ40786.1 JTDY01001036 KOB75067.1 KPJ03068.1 KQ460781 KPJ12226.1 RSAL01000010 RVE53725.1 NWSH01000400 PCG76675.1 KZ149983 PZC75749.1 KQ459606 KPI91164.1 RSAL01000291 RVE42897.1 KQ460947 KPJ10569.1 ODYU01002500 SOQ40069.1 AGBW02011630 OWR46389.1 AGBW02014435 OWR41693.1 KZ149926 PZC77547.1 NWSH01001670 PCG70462.1 ODYU01013233 SOQ59929.1 RSAL01000012 RVE53468.1 BABH01041781 RSAL01000184 RVE44850.1 PZC77545.1 ODYU01006796 SOQ48975.1 JTDY01007594 KOB65092.1 JQ814807 AFM73623.1 PZC77546.1 MF319718 ASJ26432.1 SOQ48977.1 JTDY01002277 KOB71750.1 KPI90911.1 ODYU01009953 SOQ54715.1 KQ461196 KPJ06625.1 ODYU01009427 SOQ53845.1 NWSH01007523 PCG62931.1 NWSH01003717 PCG65891.1 NEDP02003256 OWF49099.1 GEBQ01006364 JAT33613.1 AGBW02008082 OWR54177.1 GEBQ01020948 JAT19029.1 KPJ03024.1 BC082381 NEDP02005531 OWF39326.1 CM004469 OCT91179.1 ODYU01007212 SOQ49795.1 AAKN02003095 ACPB03000883 GFTR01007078 JAW09348.1 CCAG010003515 LSMT01000515 PFX16839.1 BC060482 KZ149921 PZC77721.1 JXJN01001096 CM004468 OCT93917.1 KZ150304 PZC71518.1 KQ971307 EFA11062.1 JTDY01000429 KOB77220.1 AGCU01091550 AGCU01091551 AGCU01091552
Proteomes
Interpro
Gene 3D
ProteinModelPortal
H9JHY8
A0A2H1WL24
A0A2W1BNT7
A0A194QBX2
A0A194QUH6
A0A212FPW6
+ More
A0A0L7L1G4 A0A194Q1D6 A0A0N1II42 A0A2W1B572 A0A212FBR8 H9JQ12 A0A2H1WK50 H9JQQ8 A0A2A4JBH9 A0A191XQW7 A0A3S2LBT4 A0A2H1VJ13 A0A0L7LHY6 A0A194QBZ0 A0A0N1IGL3 A0A3S2PK48 A0A2A4JXD4 A0A2W1BL36 A0A194PEP3 A0A3S2P6E3 A0A194QZ66 A0A2H1VGU5 A0A212EY15 A0A212EJL3 A0A2W1BWG2 A0A2A4JGD9 A0A2H1X3Q9 A0A3S2NZZ7 H9JWY8 A0A3S2L3L9 A0A2W1BV63 A0A2H1W795 A0A0L7KP71 I6TFN4 A0A2W1BXY3 A0A220K8L9 A0A2H1W7F1 A0A0L7L8R4 A0A194PCA8 A0A2H1WNR8 A0A194QNS4 A0A2H1WL84 A0A2A4IT83 A0A2A4J1M9 A0A210QK37 A0A1B6MCF6 A0A212FKA5 A0A1B6L5N2 A0A194QC74 Q641F6 A0A210PSA1 A0A1L8H505 P81139 A0A2H1W9L5 H0V008 T1H7M9 A0A224XHR8 A0A1B0G9P9 A0A2B4RJ44 A0A3B3ZZW1 Q6PA23 A0A2W1BTP2 A0A1B0AP02 A0A1L8HCT6 A0A3Q3WX63 A0A2W1B7Q2 D6W6G8 A0A0L7LP71 A0A1W4WIH6 K7GJB0
A0A0L7L1G4 A0A194Q1D6 A0A0N1II42 A0A2W1B572 A0A212FBR8 H9JQ12 A0A2H1WK50 H9JQQ8 A0A2A4JBH9 A0A191XQW7 A0A3S2LBT4 A0A2H1VJ13 A0A0L7LHY6 A0A194QBZ0 A0A0N1IGL3 A0A3S2PK48 A0A2A4JXD4 A0A2W1BL36 A0A194PEP3 A0A3S2P6E3 A0A194QZ66 A0A2H1VGU5 A0A212EY15 A0A212EJL3 A0A2W1BWG2 A0A2A4JGD9 A0A2H1X3Q9 A0A3S2NZZ7 H9JWY8 A0A3S2L3L9 A0A2W1BV63 A0A2H1W795 A0A0L7KP71 I6TFN4 A0A2W1BXY3 A0A220K8L9 A0A2H1W7F1 A0A0L7L8R4 A0A194PCA8 A0A2H1WNR8 A0A194QNS4 A0A2H1WL84 A0A2A4IT83 A0A2A4J1M9 A0A210QK37 A0A1B6MCF6 A0A212FKA5 A0A1B6L5N2 A0A194QC74 Q641F6 A0A210PSA1 A0A1L8H505 P81139 A0A2H1W9L5 H0V008 T1H7M9 A0A224XHR8 A0A1B0G9P9 A0A2B4RJ44 A0A3B3ZZW1 Q6PA23 A0A2W1BTP2 A0A1B0AP02 A0A1L8HCT6 A0A3Q3WX63 A0A2W1B7Q2 D6W6G8 A0A0L7LP71 A0A1W4WIH6 K7GJB0
PDB
1GPL
E-value=1.15682e-26,
Score=297
Ontologies
GO
PANTHER
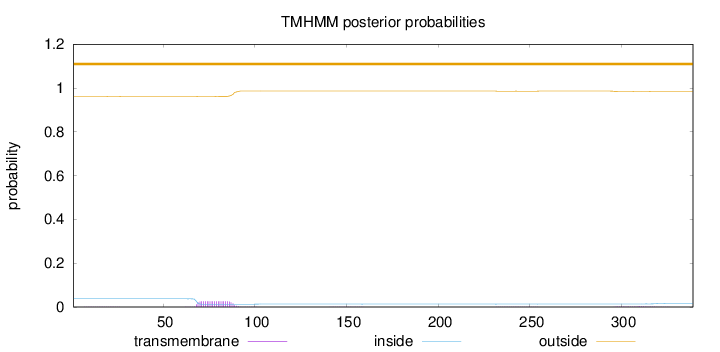
Topology
Subcellular location
Secreted
Membrane
Membrane
Length:
339
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.616369999999999
Exp number, first 60 AAs:
0.00574
Total prob of N-in:
0.03781
outside
1 - 339
Population Genetic Test Statistics
Pi
25.522971
Theta
23.597362
Tajima's D
0.298018
CLR
1.494872
CSRT
0.462926853657317
Interpretation
Uncertain
