Pre Gene Modal
BGIBMGA009138
Annotation
PREDICTED:_probable_aminopeptidase_NPEPL1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.096
Sequence
CDS
ATGTCGAATGCAACTGTAAACATAAAGTTCCAGTGGGGCTTGAAGTCCTCAGATCCCGAACAACTACCAGTTTTGGTCATAGGGCAGTCAGGACATCTGAATCAGCTTTCTTGGAGTGATGTCCGCTGTAAACTTGAGCCCAGAGTCACTGAGGAGGCATGGAAACGAGGTCTATCTTGTGTATCAACATCACCGGGTGAATCCTGTGAGCTGTGGCCTCGAGCCGTGACATTGGCCTCGTTACCAGCACGGCGCTCTCGTCATGCAGCACCTTCAAGGGCTCATGCCCTGGCCAAAATTGTTAGAGCAACTCAGCGTACTACCGATGAGGCTATAGTGCTGGTGTGCCGCAAGCGCGACGTGTACGCGAGCTGCATGGGCATCGCCCGCTCCTACCCGCTGTACCACGCGCGCTCCTCGCCGCCGCCCGCGCGCGCGCTCTCCGTCGAGATCCTGCTCGTCGCCGAGGACAAGCTCGAAGGTGAATCCGAAGACGAGGGTGTGGGACCCACAGACCCGTCGCTGCTGGACGGAGCGCTGTCCCTCGAGGAACTGACCACCATCCACCAAGCGGCCGACGCTACTCGCCTGGCCGCCAGGATCACCGACACCCCTACTAATATAATGAACGTCGATAAATTTATTGAGGAAGCGTGCAACGTGGCCAAGGAGCTGGACATCCCGGAGCCGTGCATCATCCGCGGCGAGGAGCTGCGCGCGCGCGGCATGGGCGGGCTGTACGGCGTGGGGCGCGCCGCCAGCTGCCCGCCCGCGCTCGTCACGCTGTCCTACGCCGCGCCCGCCGCCGCGCGCACCGTGGCCTGGGTCGGGAAGGGCATCGTGTACGACACCGGCGGCCTCAGCATCAAGGCGAGGGCCTCGATGGTGGGCATGAAGGGCGACTGCGGAGGCGCCGCTGCGGTGCTGGGAGCCTTCGCCCTCACCGTCAAAGCGAAGCCCGAGTTTAACCTGCACGCCGTTCTTTGCCTCGCCGAGAACTCCGTAGGACCTAACGCCACGAGGCCTGACGACGTGCACACCCTGTATTCTGGACGCACCGTCGAGATCAACAACACGGACGCCGAAGGTCGGCTGGTGCTGAGCGACGGCGTGGTGTACGCGCACCGAGACCTCAAGGCCGACACCATCGTGGACGTGGCAACACTCACCGGGGCGCAGGGCACGGCGACGGGCAAGCAGCACGCGGCGGTGCTGTGCAACGGCGCGGCGCTGGAGGCGGCGGCGGTGCGCGCGGGGCTGCGCTGCGCGGAGCTGGCGCACGCGCTGCCCTTCGCGCCCGAGCTGCACTTCGCCGAGTTCACCAGCTGCGTGGCCGACATGAAGAACAGCGTCGCGGACCGCAACAACGCGCAGCCGTCGTGCGCGGGGCTGTTCATCCTGGCGCACCTCGGGTTCGACTTCCCGGGACAGTGGCTGCACGTGGACATGGCCTCGCCCTCCGTCTGCGGCGAGCGTGCTACCGGCTACGGGGTGTCACTACTGGCCGTGCTGTTCGGCGCGCGCACCGGGTCCCGCCTGCTGCGCGCGCTTAGCCCCCTCGGTCGACTGTGA
Protein
MSNATVNIKFQWGLKSSDPEQLPVLVIGQSGHLNQLSWSDVRCKLEPRVTEEAWKRGLSCVSTSPGESCELWPRAVTLASLPARRSRHAAPSRAHALAKIVRATQRTTDEAIVLVCRKRDVYASCMGIARSYPLYHARSSPPPARALSVEILLVAEDKLEGESEDEGVGPTDPSLLDGALSLEELTTIHQAADATRLAARITDTPTNIMNVDKFIEEACNVAKELDIPEPCIIRGEELRARGMGGLYGVGRAASCPPALVTLSYAAPAAARTVAWVGKGIVYDTGGLSIKARASMVGMKGDCGGAAAVLGAFALTVKAKPEFNLHAVLCLAENSVGPNATRPDDVHTLYSGRTVEINNTDAEGRLVLSDGVVYAHRDLKADTIVDVATLTGAQGTATGKQHAAVLCNGAALEAAAVRAGLRCAELAHALPFAPELHFAEFTSCVADMKNSVADRNNAQPSCAGLFILAHLGFDFPGQWLHVDMASPSVCGERATGYGVSLLAVLFGARTGSRLLRALSPLGRL
Summary
Uniprot
H9JHZ0
A0A3S2NBG5
A0A212FPR3
A0A2H1WHS0
D6WTW2
A0A0P4W621
+ More
A0A2R5L5J7 A0A1Z5KWQ1 T1PFZ7 V5I917 A0A293LKL0 A0A1I8N7W3 A0A1I8N7W6 A0A1Y1L589 H3ASU7 A0A0K8VSZ0 A0A034W5L8 A0A0K8UT18 A0A0K8U640 A0A2J7R434 A0A0K8U2C6 A0A034W1I2 A0A0Q9XAY9 B4M3W1 B4K8J9 A0A0Q9W0Z9 W8BHX9 A0A1B0CF71 B4JEY1 A0A3P9L9L4 A0A3P9HYU8 A0A0F8CG21 A0A0M4EMY8 A0A3B3E1X2 A0A2I4CZ42 A0A0K8TL09 A0A3B3WFN4 A0A3B4WP98 A0A3B4TG61 H2MIF7 A0A1W4ZB07 A0A023GL39 T1DDR5 A0A3B4Z5P5 A0A2H4N3C8 A0A1W7RJH2 A0A1L8E2X9 A0A131XFH5 A0A087YC64 A0A0B8RRR5 A0A3B4GUX9 A0A3Q3BAH3 G3MH21 A0A3P8NNF0 A0A3P9D9E7 A9JT03 A0A146P763 A0A1B6FD08 A0A2Y9IMH1 F1Q6S1 W5MGL2 L7MDR5 A0A0S7H3Q3 A0A3P8Z2D2 A0A0R1E772 A0A1B6I8K7 A0A3B3UKW8 A0A3B5Q8H3 A0A1V4KEC8 A0A1L8EFR6 A0A067RDS8 A0A3Q2WDJ8 G3N5R0 I3KSW4 A0A3Q3L497 B4N9W9 A0A1B6JBL4 W5UBF3 A0A3Q1CMV2 A0A3P8SJR2 A0A1E1XM61 Q561T4 A0A2U9BGT4 A0A1A7YGL0 A0A151NYS5 A0A3Q1JVB9 F7DN69 A0A218V0G7 A0A1L8E2P2 F1NSZ4 A0A1A8P7D8 A0A224YNS8 A0A3B4DUE7 A0A3Q1GBX2 A0A1A8PQ66 V5IHU7 B5X304 A0A1W4UJZ3 W5KHA7
A0A2R5L5J7 A0A1Z5KWQ1 T1PFZ7 V5I917 A0A293LKL0 A0A1I8N7W3 A0A1I8N7W6 A0A1Y1L589 H3ASU7 A0A0K8VSZ0 A0A034W5L8 A0A0K8UT18 A0A0K8U640 A0A2J7R434 A0A0K8U2C6 A0A034W1I2 A0A0Q9XAY9 B4M3W1 B4K8J9 A0A0Q9W0Z9 W8BHX9 A0A1B0CF71 B4JEY1 A0A3P9L9L4 A0A3P9HYU8 A0A0F8CG21 A0A0M4EMY8 A0A3B3E1X2 A0A2I4CZ42 A0A0K8TL09 A0A3B3WFN4 A0A3B4WP98 A0A3B4TG61 H2MIF7 A0A1W4ZB07 A0A023GL39 T1DDR5 A0A3B4Z5P5 A0A2H4N3C8 A0A1W7RJH2 A0A1L8E2X9 A0A131XFH5 A0A087YC64 A0A0B8RRR5 A0A3B4GUX9 A0A3Q3BAH3 G3MH21 A0A3P8NNF0 A0A3P9D9E7 A9JT03 A0A146P763 A0A1B6FD08 A0A2Y9IMH1 F1Q6S1 W5MGL2 L7MDR5 A0A0S7H3Q3 A0A3P8Z2D2 A0A0R1E772 A0A1B6I8K7 A0A3B3UKW8 A0A3B5Q8H3 A0A1V4KEC8 A0A1L8EFR6 A0A067RDS8 A0A3Q2WDJ8 G3N5R0 I3KSW4 A0A3Q3L497 B4N9W9 A0A1B6JBL4 W5UBF3 A0A3Q1CMV2 A0A3P8SJR2 A0A1E1XM61 Q561T4 A0A2U9BGT4 A0A1A7YGL0 A0A151NYS5 A0A3Q1JVB9 F7DN69 A0A218V0G7 A0A1L8E2P2 F1NSZ4 A0A1A8P7D8 A0A224YNS8 A0A3B4DUE7 A0A3Q1GBX2 A0A1A8PQ66 V5IHU7 B5X304 A0A1W4UJZ3 W5KHA7
Pubmed
19121390
22118469
18362917
19820115
28528879
25315136
+ More
28004739 9215903 25348373 17994087 24495485 17554307 25835551 29451363 26369729 23758969 25727380 29107670 26358130 28049606 25476704 22216098 25186727 23594743 25576852 25069045 17550304 23542700 24845553 23127152 29209593 22293439 17495919 15592404 28797301 25765539 20433749 25329095
28004739 9215903 25348373 17994087 24495485 17554307 25835551 29451363 26369729 23758969 25727380 29107670 26358130 28049606 25476704 22216098 25186727 23594743 25576852 25069045 17550304 23542700 24845553 23127152 29209593 22293439 17495919 15592404 28797301 25765539 20433749 25329095
EMBL
BABH01035297
BABH01035298
RSAL01000122
RVE46662.1
AGBW02000859
OWR55748.1
+ More
ODYU01008727 SOQ52577.1 KQ971352 EFA07320.1 GDRN01069409 JAI64053.1 GGLE01000613 MBY04739.1 GFJQ02007505 JAV99464.1 KA647681 AFP62310.1 GALX01003715 JAB64751.1 GFWV01009298 MAA34027.1 GEZM01066312 GEZM01066311 JAV67968.1 AFYH01049052 AFYH01049053 GDHF01010325 JAI41989.1 GAKP01009532 JAC49420.1 GDHF01022651 JAI29663.1 GDHF01030293 JAI22021.1 NEVH01007816 PNF35586.1 GDHF01031415 JAI20899.1 GAKP01009531 JAC49421.1 CH933806 KRG00995.1 CH940652 EDW59322.1 EDW14398.2 KRF78794.1 GAMC01008193 JAB98362.1 AJWK01009751 AJWK01009752 AJWK01009753 AJWK01009754 CH916369 EDV93262.1 KQ042155 KKF18576.1 CP012526 ALC45926.1 GDAI01002772 JAI14831.1 GBBM01000817 JAC34601.1 GAAZ01000139 GBKC01000195 JAA97804.1 JAG45875.1 MG132018 ATU85537.1 GDAY02000132 JAV51281.1 GFDF01001095 JAV12989.1 GEFH01004185 JAP64396.1 AYCK01008769 GBSH01000275 JAG68749.1 JO841172 AEO32789.1 BC155148 AAI55149.1 GCES01146548 JAQ39774.1 GECZ01021775 JAS47994.1 CU467627 CU467883 AHAT01024342 AHAT01024343 GACK01003067 JAA61967.1 GBYX01445620 JAO35813.1 CM000160 KRK03554.1 GECU01024441 JAS83265.1 LSYS01003456 OPJ82745.1 GFDG01001293 JAV17506.1 KK852570 KDR21148.1 AERX01006798 CH964232 EDW81724.1 GECU01011046 JAS96660.1 JT410979 AHH39258.1 GFAA01003090 JAU00345.1 BC093321 AAH93321.1 CP026248 AWP03029.1 HADW01001351 HADX01007139 SBP29371.1 AKHW03001485 KYO42031.1 MUZQ01000085 OWK59112.1 GFDF01001096 JAV12988.1 AADN05000479 HAEI01001676 HAEH01016756 SBR77191.1 GFPF01004366 MAA15512.1 HAEF01009391 HAEG01009015 SBR83393.1 GANP01004317 JAB80151.1 BT045423 ACI33685.1
ODYU01008727 SOQ52577.1 KQ971352 EFA07320.1 GDRN01069409 JAI64053.1 GGLE01000613 MBY04739.1 GFJQ02007505 JAV99464.1 KA647681 AFP62310.1 GALX01003715 JAB64751.1 GFWV01009298 MAA34027.1 GEZM01066312 GEZM01066311 JAV67968.1 AFYH01049052 AFYH01049053 GDHF01010325 JAI41989.1 GAKP01009532 JAC49420.1 GDHF01022651 JAI29663.1 GDHF01030293 JAI22021.1 NEVH01007816 PNF35586.1 GDHF01031415 JAI20899.1 GAKP01009531 JAC49421.1 CH933806 KRG00995.1 CH940652 EDW59322.1 EDW14398.2 KRF78794.1 GAMC01008193 JAB98362.1 AJWK01009751 AJWK01009752 AJWK01009753 AJWK01009754 CH916369 EDV93262.1 KQ042155 KKF18576.1 CP012526 ALC45926.1 GDAI01002772 JAI14831.1 GBBM01000817 JAC34601.1 GAAZ01000139 GBKC01000195 JAA97804.1 JAG45875.1 MG132018 ATU85537.1 GDAY02000132 JAV51281.1 GFDF01001095 JAV12989.1 GEFH01004185 JAP64396.1 AYCK01008769 GBSH01000275 JAG68749.1 JO841172 AEO32789.1 BC155148 AAI55149.1 GCES01146548 JAQ39774.1 GECZ01021775 JAS47994.1 CU467627 CU467883 AHAT01024342 AHAT01024343 GACK01003067 JAA61967.1 GBYX01445620 JAO35813.1 CM000160 KRK03554.1 GECU01024441 JAS83265.1 LSYS01003456 OPJ82745.1 GFDG01001293 JAV17506.1 KK852570 KDR21148.1 AERX01006798 CH964232 EDW81724.1 GECU01011046 JAS96660.1 JT410979 AHH39258.1 GFAA01003090 JAU00345.1 BC093321 AAH93321.1 CP026248 AWP03029.1 HADW01001351 HADX01007139 SBP29371.1 AKHW03001485 KYO42031.1 MUZQ01000085 OWK59112.1 GFDF01001096 JAV12988.1 AADN05000479 HAEI01001676 HAEH01016756 SBR77191.1 GFPF01004366 MAA15512.1 HAEF01009391 HAEG01009015 SBR83393.1 GANP01004317 JAB80151.1 BT045423 ACI33685.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000007266
UP000095301
UP000008672
+ More
UP000235965 UP000009192 UP000008792 UP000092461 UP000001070 UP000265180 UP000265200 UP000092553 UP000261560 UP000192220 UP000261480 UP000261360 UP000261420 UP000001038 UP000192224 UP000261400 UP000028760 UP000261460 UP000264800 UP000265100 UP000265160 UP000248482 UP000000437 UP000018468 UP000265140 UP000002282 UP000261500 UP000002852 UP000190648 UP000027135 UP000264840 UP000007635 UP000005207 UP000261640 UP000007798 UP000221080 UP000257160 UP000265080 UP000246464 UP000050525 UP000265040 UP000002280 UP000197619 UP000000539 UP000261440 UP000257200 UP000087266 UP000192221 UP000018467
UP000235965 UP000009192 UP000008792 UP000092461 UP000001070 UP000265180 UP000265200 UP000092553 UP000261560 UP000192220 UP000261480 UP000261360 UP000261420 UP000001038 UP000192224 UP000261400 UP000028760 UP000261460 UP000264800 UP000265100 UP000265160 UP000248482 UP000000437 UP000018468 UP000265140 UP000002282 UP000261500 UP000002852 UP000190648 UP000027135 UP000264840 UP000007635 UP000005207 UP000261640 UP000007798 UP000221080 UP000257160 UP000265080 UP000246464 UP000050525 UP000265040 UP000002280 UP000197619 UP000000539 UP000261440 UP000257200 UP000087266 UP000192221 UP000018467
Interpro
SUPFAM
SSF55186
SSF55186
ProteinModelPortal
H9JHZ0
A0A3S2NBG5
A0A212FPR3
A0A2H1WHS0
D6WTW2
A0A0P4W621
+ More
A0A2R5L5J7 A0A1Z5KWQ1 T1PFZ7 V5I917 A0A293LKL0 A0A1I8N7W3 A0A1I8N7W6 A0A1Y1L589 H3ASU7 A0A0K8VSZ0 A0A034W5L8 A0A0K8UT18 A0A0K8U640 A0A2J7R434 A0A0K8U2C6 A0A034W1I2 A0A0Q9XAY9 B4M3W1 B4K8J9 A0A0Q9W0Z9 W8BHX9 A0A1B0CF71 B4JEY1 A0A3P9L9L4 A0A3P9HYU8 A0A0F8CG21 A0A0M4EMY8 A0A3B3E1X2 A0A2I4CZ42 A0A0K8TL09 A0A3B3WFN4 A0A3B4WP98 A0A3B4TG61 H2MIF7 A0A1W4ZB07 A0A023GL39 T1DDR5 A0A3B4Z5P5 A0A2H4N3C8 A0A1W7RJH2 A0A1L8E2X9 A0A131XFH5 A0A087YC64 A0A0B8RRR5 A0A3B4GUX9 A0A3Q3BAH3 G3MH21 A0A3P8NNF0 A0A3P9D9E7 A9JT03 A0A146P763 A0A1B6FD08 A0A2Y9IMH1 F1Q6S1 W5MGL2 L7MDR5 A0A0S7H3Q3 A0A3P8Z2D2 A0A0R1E772 A0A1B6I8K7 A0A3B3UKW8 A0A3B5Q8H3 A0A1V4KEC8 A0A1L8EFR6 A0A067RDS8 A0A3Q2WDJ8 G3N5R0 I3KSW4 A0A3Q3L497 B4N9W9 A0A1B6JBL4 W5UBF3 A0A3Q1CMV2 A0A3P8SJR2 A0A1E1XM61 Q561T4 A0A2U9BGT4 A0A1A7YGL0 A0A151NYS5 A0A3Q1JVB9 F7DN69 A0A218V0G7 A0A1L8E2P2 F1NSZ4 A0A1A8P7D8 A0A224YNS8 A0A3B4DUE7 A0A3Q1GBX2 A0A1A8PQ66 V5IHU7 B5X304 A0A1W4UJZ3 W5KHA7
A0A2R5L5J7 A0A1Z5KWQ1 T1PFZ7 V5I917 A0A293LKL0 A0A1I8N7W3 A0A1I8N7W6 A0A1Y1L589 H3ASU7 A0A0K8VSZ0 A0A034W5L8 A0A0K8UT18 A0A0K8U640 A0A2J7R434 A0A0K8U2C6 A0A034W1I2 A0A0Q9XAY9 B4M3W1 B4K8J9 A0A0Q9W0Z9 W8BHX9 A0A1B0CF71 B4JEY1 A0A3P9L9L4 A0A3P9HYU8 A0A0F8CG21 A0A0M4EMY8 A0A3B3E1X2 A0A2I4CZ42 A0A0K8TL09 A0A3B3WFN4 A0A3B4WP98 A0A3B4TG61 H2MIF7 A0A1W4ZB07 A0A023GL39 T1DDR5 A0A3B4Z5P5 A0A2H4N3C8 A0A1W7RJH2 A0A1L8E2X9 A0A131XFH5 A0A087YC64 A0A0B8RRR5 A0A3B4GUX9 A0A3Q3BAH3 G3MH21 A0A3P8NNF0 A0A3P9D9E7 A9JT03 A0A146P763 A0A1B6FD08 A0A2Y9IMH1 F1Q6S1 W5MGL2 L7MDR5 A0A0S7H3Q3 A0A3P8Z2D2 A0A0R1E772 A0A1B6I8K7 A0A3B3UKW8 A0A3B5Q8H3 A0A1V4KEC8 A0A1L8EFR6 A0A067RDS8 A0A3Q2WDJ8 G3N5R0 I3KSW4 A0A3Q3L497 B4N9W9 A0A1B6JBL4 W5UBF3 A0A3Q1CMV2 A0A3P8SJR2 A0A1E1XM61 Q561T4 A0A2U9BGT4 A0A1A7YGL0 A0A151NYS5 A0A3Q1JVB9 F7DN69 A0A218V0G7 A0A1L8E2P2 F1NSZ4 A0A1A8P7D8 A0A224YNS8 A0A3B4DUE7 A0A3Q1GBX2 A0A1A8PQ66 V5IHU7 B5X304 A0A1W4UJZ3 W5KHA7
PDB
2HC9
E-value=1.15241e-64,
Score=627
Ontologies
GO
PANTHER
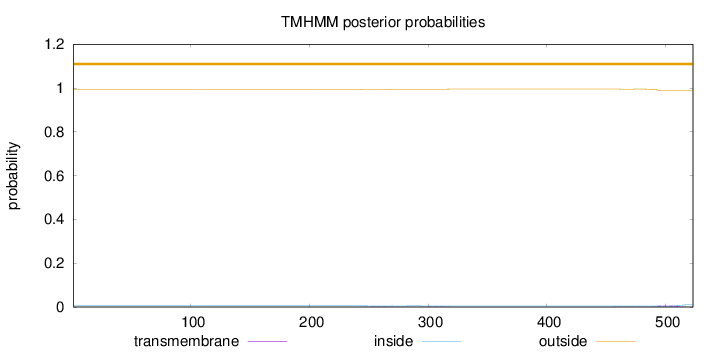
Topology
Length:
523
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.20835
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00582
outside
1 - 523
Population Genetic Test Statistics
Pi
27.04194
Theta
26.088721
Tajima's D
-0.833539
CLR
0.393045
CSRT
0.173991300434978
Interpretation
Uncertain
