Pre Gene Modal
BGIBMGA012520
Annotation
TP53-regulating_kinase_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 1.973
Sequence
CDS
ATGACTGAAGATTTGAAGATATTAAAACAAGGAGCTGAAGCAAAATTGTACATTTGCAATTATCTTGGAAGACCTACATTGATCAAGGAAAGGTTCGTTAAAAATTACAGACATCTTGATTTAGATACAAATATAACAAAAGAGAGAATTAAAAATGAAGCACGTTCCATTGTACGCTGTAAAACAGCCGGAATTCGTACACCGGCTTTATATTTAGTTGATTTTGAGCGTCGTCGAATTTACATGGAACATTTCGAACGGAACGTAACTGTCAAAGATTTTATCATAAATTTGACGAAAACTGGCAGTGAAAATGAAACAAACAGCCTGAACATACTAGCCAAAATGATTGGCGAAACTGTTCGAACATTACACGACACCAACATTATACACGGAGACTTGACAACTTCTAATATATTGTTGATACCGAAAAATGACAATGATAATTGGATTCAATGCTGTGAATTTCATTTGGTCATGATTGATTTTGGTTTATCATATGTTGACTCTAGTGCCGAAGACAAAGGCGTTGACCTATATGTCCTAGAAAGGGCATTAATAAGTACCCACAATGACTATCCAGAGTTATTCAATAAAATGTATGATGCATACAAAAGTTCTAGTAAGAGCAATATAAAAGAAATCATCAGTAAATATGAAGAAGTCAGAGCTAGAGGGAGAAAGAGAACTATGGTTGGTTGA
Protein
MTEDLKILKQGAEAKLYICNYLGRPTLIKERFVKNYRHLDLDTNITKERIKNEARSIVRCKTAGIRTPALYLVDFERRRIYMEHFERNVTVKDFIINLTKTGSENETNSLNILAKMIGETVRTLHDTNIIHGDLTTSNILLIPKNDNDNWIQCCEFHLVMIDFGLSYVDSSAEDKGVDLYVLERALISTHNDYPELFNKMYDAYKSSSKSNIKEIISKYEEVRARGRKRTMVG
Summary
Uniprot
H9JSK9
S4PE12
A0A0L7L9J3
A0A194QTI6
A0A2W1BSX6
A0A2A4J7C1
+ More
A0A194QDM2 A0A212FPS3 A0A2H1W2F8 D6WFA1 A0A2J7R8E9 A0A2P8YE81 A0A067QP37 A0A1Y1LQ36 J3JW11 N6T5X8 A0A023F274 A0A182XYW0 A0A084WN99 A0A1I8NQG3 A0A336KB71 A0A3L8D932 A0A0V0GAT5 A0A182PQF4 E2AID2 A0A1J1J7U8 Q7QJE5 A0A182X3R5 A0A026WWJ3 A0A151JSJ8 A0A182I3A4 A0A182U0S5 A0A1S3H2B9 F4WFX5 A0A195BH27 A0A0K8UP22 A0A195F0I3 A0A158NL40 A0A182WJ86 A0A2M3ZJN7 A0A182KRC9 K7J5V6 T1I1Q1 W5JNL0 A0A2M4BZL1 E2BKH6 A0A1I8NHE6 A0A1B6DMU1 A0A034VGH9 A0A182MSM6 A0A182V704 A0A151XBJ6 A0A2M4BYL3 B4MXF3 A0A0B6ZE33 A0A1A9VN21 A0A310SC88 A0A232F5D5 A0A0N0BIF4 A0A0M4EFM2 A0A2A3EK03 R7UIC9 A0A182QFX2 A0A087ZZD9 A0A182KGH7 A0A2M4BZ29 A0A1W4US45 A0A1B6H149 A0A0P4VUA6 B4QRI3 B4HUD6 B3M405 A0A0L7QNF7 Q4V7S0 A0A1Q3F915 A0A023EKP4 B4IX00 A0A182G5T4 V9LAM3 Q176Z1 A0A154PK25 A0A2T7NI99 A0A2M4AY46 A0A182RSN2 A0A1S4FD31 B4PIE2 A0A3B3C8Q0 A0A1L8DLY2 Q9VRJ6 A0A1L8DTJ3 A0A0C9R7Y2 A0A195BY27 A0A267FHV5 K1R079 A0A182JHA5 A9JTT7 F6ZWY1 A0A3P8TJD8 A0A3N0XUP7 A0A267FGA2
A0A194QDM2 A0A212FPS3 A0A2H1W2F8 D6WFA1 A0A2J7R8E9 A0A2P8YE81 A0A067QP37 A0A1Y1LQ36 J3JW11 N6T5X8 A0A023F274 A0A182XYW0 A0A084WN99 A0A1I8NQG3 A0A336KB71 A0A3L8D932 A0A0V0GAT5 A0A182PQF4 E2AID2 A0A1J1J7U8 Q7QJE5 A0A182X3R5 A0A026WWJ3 A0A151JSJ8 A0A182I3A4 A0A182U0S5 A0A1S3H2B9 F4WFX5 A0A195BH27 A0A0K8UP22 A0A195F0I3 A0A158NL40 A0A182WJ86 A0A2M3ZJN7 A0A182KRC9 K7J5V6 T1I1Q1 W5JNL0 A0A2M4BZL1 E2BKH6 A0A1I8NHE6 A0A1B6DMU1 A0A034VGH9 A0A182MSM6 A0A182V704 A0A151XBJ6 A0A2M4BYL3 B4MXF3 A0A0B6ZE33 A0A1A9VN21 A0A310SC88 A0A232F5D5 A0A0N0BIF4 A0A0M4EFM2 A0A2A3EK03 R7UIC9 A0A182QFX2 A0A087ZZD9 A0A182KGH7 A0A2M4BZ29 A0A1W4US45 A0A1B6H149 A0A0P4VUA6 B4QRI3 B4HUD6 B3M405 A0A0L7QNF7 Q4V7S0 A0A1Q3F915 A0A023EKP4 B4IX00 A0A182G5T4 V9LAM3 Q176Z1 A0A154PK25 A0A2T7NI99 A0A2M4AY46 A0A182RSN2 A0A1S4FD31 B4PIE2 A0A3B3C8Q0 A0A1L8DLY2 Q9VRJ6 A0A1L8DTJ3 A0A0C9R7Y2 A0A195BY27 A0A267FHV5 K1R079 A0A182JHA5 A9JTT7 F6ZWY1 A0A3P8TJD8 A0A3N0XUP7 A0A267FGA2
Pubmed
19121390
23622113
26227816
26354079
28756777
22118469
+ More
18362917 19820115 29403074 24845553 28004739 22516182 23537049 25474469 25244985 24438588 30249741 20798317 12364791 14747013 17210077 24508170 21719571 21347285 20966253 20075255 20920257 23761445 25315136 25348373 17994087 28648823 23254933 27129103 22936249 24945155 26483478 24402279 17510324 17550304 29451363 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22992520 20431018
18362917 19820115 29403074 24845553 28004739 22516182 23537049 25474469 25244985 24438588 30249741 20798317 12364791 14747013 17210077 24508170 21719571 21347285 20966253 20075255 20920257 23761445 25315136 25348373 17994087 28648823 23254933 27129103 22936249 24945155 26483478 24402279 17510324 17550304 29451363 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22992520 20431018
EMBL
BABH01041200
GAIX01003396
JAA89164.1
JTDY01002118
KOB72065.1
KQ461154
+ More
KPJ08624.1 KZ149947 PZC76735.1 NWSH01002907 PCG67322.1 KQ459193 KPJ03060.1 AGBW02000859 OWR55752.1 ODYU01005877 SOQ47217.1 KQ971319 EFA00476.1 NEVH01006722 PNF37098.1 PYGN01000666 PSN42562.1 KK853119 KDR11130.1 GEZM01051279 GEZM01051277 JAV75061.1 BT127429 AEE62391.1 APGK01043155 KB741011 ENN75584.1 GBBI01003526 JAC15186.1 ATLV01024580 KE525352 KFB51693.1 UFQS01000164 UFQT01000164 SSX00675.1 SSX21055.1 QOIP01000011 RLU16816.1 GECL01000874 JAP05250.1 GL439757 EFN66807.1 CVRI01000069 CRL06977.1 AAAB01008807 EAA04620.4 KK107078 EZA60422.1 KQ978530 KYN30326.1 APCN01000204 GL888128 EGI66742.1 KQ976488 KYM83533.1 GDHF01023865 JAI28449.1 KQ981866 KYN34090.1 ADTU01019238 ADTU01019239 GGFM01007907 MBW28658.1 ACPB03014712 ADMH02000549 ETN65952.1 GGFJ01009027 MBW58168.1 GL448795 EFN83816.1 GEDC01010348 JAS26950.1 GAKP01018302 GAKP01018299 JAC40653.1 AXCM01005343 KQ982320 KYQ57741.1 GGFJ01009026 MBW58167.1 CH963876 EDW76722.1 HACG01019075 CEK65940.1 KQ779356 OAD51967.1 NNAY01000999 OXU25547.1 KQ435732 KOX77364.1 CP012525 ALC43077.1 KZ288230 PBC31592.1 AMQN01007664 KB301152 ELU05858.1 AXCN02000379 GGFJ01009142 MBW58283.1 GECZ01001367 JAS68402.1 GDKW01000342 JAI56253.1 CM000363 CM002912 EDX09294.1 KMY97705.1 CH480817 EDW50557.1 CH902618 EDV40367.1 KQ414855 KOC60153.1 BC097750 AAH97750.1 GFDL01010981 JAV24064.1 JXUM01020768 GAPW01003871 KQ560581 JAC09727.1 KXJ81810.1 CH916366 EDV96306.1 JXUM01043910 KQ561380 KXJ78775.1 JW876233 AFP08750.1 CH477380 EAT42218.1 KQ434938 KZC12147.1 PZQS01000012 PVD20899.1 GGFK01012379 MBW45700.1 CM000159 EDW93484.1 GFDF01006707 JAV07377.1 AE014296 BT022659 AAF50799.1 AAY55075.1 GFDF01004343 JAV09741.1 GBYB01004140 JAG73907.1 KQ978501 KYM93497.1 NIVC01001064 PAA72727.1 JH818091 EKC36864.1 BC155471 AAI55472.1 AAMC01020205 RJVU01060476 ROJ48030.1 NIVC01001063 PAA72743.1
KPJ08624.1 KZ149947 PZC76735.1 NWSH01002907 PCG67322.1 KQ459193 KPJ03060.1 AGBW02000859 OWR55752.1 ODYU01005877 SOQ47217.1 KQ971319 EFA00476.1 NEVH01006722 PNF37098.1 PYGN01000666 PSN42562.1 KK853119 KDR11130.1 GEZM01051279 GEZM01051277 JAV75061.1 BT127429 AEE62391.1 APGK01043155 KB741011 ENN75584.1 GBBI01003526 JAC15186.1 ATLV01024580 KE525352 KFB51693.1 UFQS01000164 UFQT01000164 SSX00675.1 SSX21055.1 QOIP01000011 RLU16816.1 GECL01000874 JAP05250.1 GL439757 EFN66807.1 CVRI01000069 CRL06977.1 AAAB01008807 EAA04620.4 KK107078 EZA60422.1 KQ978530 KYN30326.1 APCN01000204 GL888128 EGI66742.1 KQ976488 KYM83533.1 GDHF01023865 JAI28449.1 KQ981866 KYN34090.1 ADTU01019238 ADTU01019239 GGFM01007907 MBW28658.1 ACPB03014712 ADMH02000549 ETN65952.1 GGFJ01009027 MBW58168.1 GL448795 EFN83816.1 GEDC01010348 JAS26950.1 GAKP01018302 GAKP01018299 JAC40653.1 AXCM01005343 KQ982320 KYQ57741.1 GGFJ01009026 MBW58167.1 CH963876 EDW76722.1 HACG01019075 CEK65940.1 KQ779356 OAD51967.1 NNAY01000999 OXU25547.1 KQ435732 KOX77364.1 CP012525 ALC43077.1 KZ288230 PBC31592.1 AMQN01007664 KB301152 ELU05858.1 AXCN02000379 GGFJ01009142 MBW58283.1 GECZ01001367 JAS68402.1 GDKW01000342 JAI56253.1 CM000363 CM002912 EDX09294.1 KMY97705.1 CH480817 EDW50557.1 CH902618 EDV40367.1 KQ414855 KOC60153.1 BC097750 AAH97750.1 GFDL01010981 JAV24064.1 JXUM01020768 GAPW01003871 KQ560581 JAC09727.1 KXJ81810.1 CH916366 EDV96306.1 JXUM01043910 KQ561380 KXJ78775.1 JW876233 AFP08750.1 CH477380 EAT42218.1 KQ434938 KZC12147.1 PZQS01000012 PVD20899.1 GGFK01012379 MBW45700.1 CM000159 EDW93484.1 GFDF01006707 JAV07377.1 AE014296 BT022659 AAF50799.1 AAY55075.1 GFDF01004343 JAV09741.1 GBYB01004140 JAG73907.1 KQ978501 KYM93497.1 NIVC01001064 PAA72727.1 JH818091 EKC36864.1 BC155471 AAI55472.1 AAMC01020205 RJVU01060476 ROJ48030.1 NIVC01001063 PAA72743.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000218220
UP000053268
UP000007151
+ More
UP000007266 UP000235965 UP000245037 UP000027135 UP000019118 UP000076408 UP000030765 UP000095300 UP000279307 UP000075885 UP000000311 UP000183832 UP000007062 UP000076407 UP000053097 UP000078492 UP000075840 UP000075902 UP000085678 UP000007755 UP000078540 UP000078541 UP000005205 UP000075920 UP000075882 UP000002358 UP000015103 UP000000673 UP000008237 UP000095301 UP000075883 UP000075903 UP000075809 UP000007798 UP000078200 UP000215335 UP000053105 UP000092553 UP000242457 UP000014760 UP000075886 UP000005203 UP000075881 UP000192221 UP000000304 UP000001292 UP000007801 UP000053825 UP000069940 UP000249989 UP000001070 UP000008820 UP000076502 UP000245119 UP000075900 UP000002282 UP000261560 UP000000803 UP000078542 UP000215902 UP000005408 UP000075880 UP000008143 UP000265080
UP000007266 UP000235965 UP000245037 UP000027135 UP000019118 UP000076408 UP000030765 UP000095300 UP000279307 UP000075885 UP000000311 UP000183832 UP000007062 UP000076407 UP000053097 UP000078492 UP000075840 UP000075902 UP000085678 UP000007755 UP000078540 UP000078541 UP000005205 UP000075920 UP000075882 UP000002358 UP000015103 UP000000673 UP000008237 UP000095301 UP000075883 UP000075903 UP000075809 UP000007798 UP000078200 UP000215335 UP000053105 UP000092553 UP000242457 UP000014760 UP000075886 UP000005203 UP000075881 UP000192221 UP000000304 UP000001292 UP000007801 UP000053825 UP000069940 UP000249989 UP000001070 UP000008820 UP000076502 UP000245119 UP000075900 UP000002282 UP000261560 UP000000803 UP000078542 UP000215902 UP000005408 UP000075880 UP000008143 UP000265080
Interpro
IPR008266
Tyr_kinase_AS
+ More
IPR000719 Prot_kinase_dom
IPR022495 Bud32
IPR011009 Kinase-like_dom_sf
IPR012675 Beta-grasp_dom_sf
IPR018163 Thr/Ala-tRNA-synth_IIc_edit
IPR012947 tRNA_SAD
IPR003593 AAA+_ATPase
IPR003959 ATPase_AAA_core
IPR013748 Rep_factorC_C
IPR027417 P-loop_NTPase
IPR008921 DNA_pol3_clamp-load_cplx_C
IPR000719 Prot_kinase_dom
IPR022495 Bud32
IPR011009 Kinase-like_dom_sf
IPR012675 Beta-grasp_dom_sf
IPR018163 Thr/Ala-tRNA-synth_IIc_edit
IPR012947 tRNA_SAD
IPR003593 AAA+_ATPase
IPR003959 ATPase_AAA_core
IPR013748 Rep_factorC_C
IPR027417 P-loop_NTPase
IPR008921 DNA_pol3_clamp-load_cplx_C
Gene 3D
ProteinModelPortal
H9JSK9
S4PE12
A0A0L7L9J3
A0A194QTI6
A0A2W1BSX6
A0A2A4J7C1
+ More
A0A194QDM2 A0A212FPS3 A0A2H1W2F8 D6WFA1 A0A2J7R8E9 A0A2P8YE81 A0A067QP37 A0A1Y1LQ36 J3JW11 N6T5X8 A0A023F274 A0A182XYW0 A0A084WN99 A0A1I8NQG3 A0A336KB71 A0A3L8D932 A0A0V0GAT5 A0A182PQF4 E2AID2 A0A1J1J7U8 Q7QJE5 A0A182X3R5 A0A026WWJ3 A0A151JSJ8 A0A182I3A4 A0A182U0S5 A0A1S3H2B9 F4WFX5 A0A195BH27 A0A0K8UP22 A0A195F0I3 A0A158NL40 A0A182WJ86 A0A2M3ZJN7 A0A182KRC9 K7J5V6 T1I1Q1 W5JNL0 A0A2M4BZL1 E2BKH6 A0A1I8NHE6 A0A1B6DMU1 A0A034VGH9 A0A182MSM6 A0A182V704 A0A151XBJ6 A0A2M4BYL3 B4MXF3 A0A0B6ZE33 A0A1A9VN21 A0A310SC88 A0A232F5D5 A0A0N0BIF4 A0A0M4EFM2 A0A2A3EK03 R7UIC9 A0A182QFX2 A0A087ZZD9 A0A182KGH7 A0A2M4BZ29 A0A1W4US45 A0A1B6H149 A0A0P4VUA6 B4QRI3 B4HUD6 B3M405 A0A0L7QNF7 Q4V7S0 A0A1Q3F915 A0A023EKP4 B4IX00 A0A182G5T4 V9LAM3 Q176Z1 A0A154PK25 A0A2T7NI99 A0A2M4AY46 A0A182RSN2 A0A1S4FD31 B4PIE2 A0A3B3C8Q0 A0A1L8DLY2 Q9VRJ6 A0A1L8DTJ3 A0A0C9R7Y2 A0A195BY27 A0A267FHV5 K1R079 A0A182JHA5 A9JTT7 F6ZWY1 A0A3P8TJD8 A0A3N0XUP7 A0A267FGA2
A0A194QDM2 A0A212FPS3 A0A2H1W2F8 D6WFA1 A0A2J7R8E9 A0A2P8YE81 A0A067QP37 A0A1Y1LQ36 J3JW11 N6T5X8 A0A023F274 A0A182XYW0 A0A084WN99 A0A1I8NQG3 A0A336KB71 A0A3L8D932 A0A0V0GAT5 A0A182PQF4 E2AID2 A0A1J1J7U8 Q7QJE5 A0A182X3R5 A0A026WWJ3 A0A151JSJ8 A0A182I3A4 A0A182U0S5 A0A1S3H2B9 F4WFX5 A0A195BH27 A0A0K8UP22 A0A195F0I3 A0A158NL40 A0A182WJ86 A0A2M3ZJN7 A0A182KRC9 K7J5V6 T1I1Q1 W5JNL0 A0A2M4BZL1 E2BKH6 A0A1I8NHE6 A0A1B6DMU1 A0A034VGH9 A0A182MSM6 A0A182V704 A0A151XBJ6 A0A2M4BYL3 B4MXF3 A0A0B6ZE33 A0A1A9VN21 A0A310SC88 A0A232F5D5 A0A0N0BIF4 A0A0M4EFM2 A0A2A3EK03 R7UIC9 A0A182QFX2 A0A087ZZD9 A0A182KGH7 A0A2M4BZ29 A0A1W4US45 A0A1B6H149 A0A0P4VUA6 B4QRI3 B4HUD6 B3M405 A0A0L7QNF7 Q4V7S0 A0A1Q3F915 A0A023EKP4 B4IX00 A0A182G5T4 V9LAM3 Q176Z1 A0A154PK25 A0A2T7NI99 A0A2M4AY46 A0A182RSN2 A0A1S4FD31 B4PIE2 A0A3B3C8Q0 A0A1L8DLY2 Q9VRJ6 A0A1L8DTJ3 A0A0C9R7Y2 A0A195BY27 A0A267FHV5 K1R079 A0A182JHA5 A9JTT7 F6ZWY1 A0A3P8TJD8 A0A3N0XUP7 A0A267FGA2
PDB
4WWA
E-value=5.29158e-22,
Score=255
Ontologies
GO
PANTHER
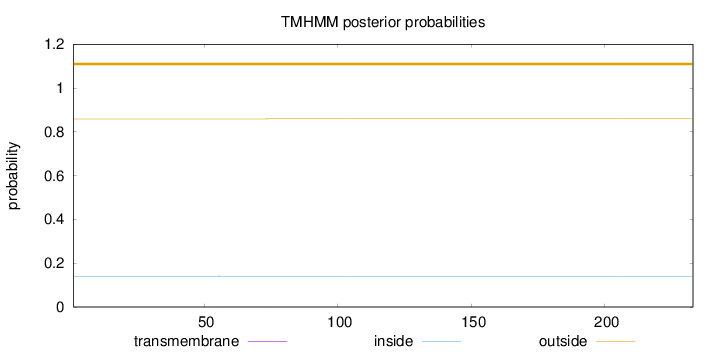
Topology
Length:
233
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00436
Exp number, first 60 AAs:
0.00153
Total prob of N-in:
0.14084
outside
1 - 233
Population Genetic Test Statistics
Pi
320.985703
Theta
227.627215
Tajima's D
1.179933
CLR
0.257724
CSRT
0.707114644267787
Interpretation
Uncertain
