Gene
KWMTBOMO12259
Pre Gene Modal
BGIBMGA009144
Annotation
PREDICTED:_gastrulation_defective_protein_1_homolog_[Papilio_xuthus]
Full name
Gastrulation defective protein 1 homolog
Location in the cell
Nuclear Reliability : 4.742
Sequence
CDS
ATGAGTAAAAAGCCCATATCGTTCGGTAAAATAAGTCTAAATTTTACTAAAGCAAAGGAATTTAATGATAAAAATGAAAAGAAATCGCCAGCAGGATTTGGAACCTTTGGTCGAACTCCTATACAAGAACAAAAAGAAATTGAAGAAATCGCCGACGATTTAGAGAGCCAGCACGTAGAACAAGTCATGGGCATTAAAAACTTTGGTAAAAAAGCCAAAAATTTCAATTTAGATGAGTTAGTTGAGCAAGCAAAGAAAACTGCACAAGAAGCCAACAAAAAGAGGTTGGAAGCGGAATTGAATGCTGATAGAAATTCATCTTCTGATGACCAGGTTGGCAATACTATTGGACCCACATTGGCCAATGTTGACCAAAATCCTTCAAAAGATGATGACGAAGGATTAATAGGACCTTTACCGCCGATGTCTCCGGACGCGAGTAAAGAACAAGAAGAAGATAAAGAGGAGGATAGCTATGACGAGCTGAGTGGTTCTGATGATGAAGAAATAAGTTTGGTAAACAGAATACCAAATACACATGAGGTAGAAATGCATCATGGTTCAAAAGCAGTGATAGCACTTGCTGCCGATCCTTCTGGAGCTAGACTAGCCACTGGATCCATTGATTACGATGTATCCTTCTGGGATTTTGCAGGTATGGACTCGTCAATGCGTTCGTTCCGCACTCTTCAGCCCTGCGAGAACCATCCTATCAAAGCCCTGCAGTACTCAGCTACAGGAGACTCGATCCTCGTGGTCAGCGGTTCAGCACAAGCTAAAGTACTAGACAGAGACGGTTTTGAAGTCTTGGAATGCGTGAAAGGAGATCAGTATATCACGGATATGGCAAAGACGAAAGGTCACACGGCGTCCTTGAACAGCGGCTGCTGGCACCCTCACATCCGCGAGGAGTTCATGACCTGCTCTCAAGATGGAACGCTCAGGCTCTGGCTAACGGAAAATCCGAAACAACAGAAATCCGTCATCAAACCCCGTCAGCAAGGCGGCTTGAAGACCAACCCCACCGCCTGCACATTCTCCAGAGACGGTAACACTGTAGCGTGCGGCTGTTTCGATGGCTCCATCCAGATGTGGGACCACAGGAAGAACTTCGTCAACACCACGACCTTGCTCAGGGACGCTCATCAGAAACAGACGGAGATCACGAGCATCGCGTACTCATACTTAAGGCAACACCTGGCCACTCGCAGCAACGATGATACGCTAAAACTGTGGGACTTGAGGAACTTCAAGAAACCCATCAAAGTGTTTGCGGACTTGTTCTCGAGATACGAACAGACCGATTGCGCCTTCAGCCCAGACGACTCTATGGTGTTCACGGGCGAGTCTCTCCAGCGTGGCCAAGAGGTGGGCAAATTGATATTCTGGGACACGAAAACTTTCGAACAGGTCAACACAATTGAGGTCAGTTCGTCCCACGTCATCAAAGCGATGTGGCACCCGAAATTGAATCAGATATTCGTCGGATGCGGCAACGGCGTCGTGAAATGCTATTACGATAAAAAGAGAAGTCTCCGCGGCGCCAAGCTCTGTATAATAAAGACGCACAGGAAAAAGGACACTATTGAGGTGGTGAGCACACAGCAGATCATAACCCCGCACGCTTTGCCCCTGTTCAGACAAGAAAAACTTCGGACCAGCAAGAAAAAGATGGAAAAAGAGAGGCTGGACCCGGTTAAGTCACACAGACCAGACCTGCCCATAACTTCAGGGCAGGGTGGGAGAGTAGCTGCTTCGGGTAGCACTCTGAGCTCATTCGTCATCAGGAATTTGGGGCTGAGTAAGAGAGTTAACGATGAACAGGACCCGAGGGAGGCCATATTGAAATATGCTAAAGATGCAGAAGAGAATCCCTTCTGGGTAGCTCCGGCTTATAAGAAAACTCAGCCGAAACCTATCTTCCAAGGGTCTGACGAGGAAGGACCGTCCGATCCTAAGAAGTCGAAGACTTCGTGA
Protein
MSKKPISFGKISLNFTKAKEFNDKNEKKSPAGFGTFGRTPIQEQKEIEEIADDLESQHVEQVMGIKNFGKKAKNFNLDELVEQAKKTAQEANKKRLEAELNADRNSSSDDQVGNTIGPTLANVDQNPSKDDDEGLIGPLPPMSPDASKEQEEDKEEDSYDELSGSDDEEISLVNRIPNTHEVEMHHGSKAVIALAADPSGARLATGSIDYDVSFWDFAGMDSSMRSFRTLQPCENHPIKALQYSATGDSILVVSGSAQAKVLDRDGFEVLECVKGDQYITDMAKTKGHTASLNSGCWHPHIREEFMTCSQDGTLRLWLTENPKQQKSVIKPRQQGGLKTNPTACTFSRDGNTVACGCFDGSIQMWDHRKNFVNTTTLLRDAHQKQTEITSIAYSYLRQHLATRSNDDTLKLWDLRNFKKPIKVFADLFSRYEQTDCAFSPDDSMVFTGESLQRGQEVGKLIFWDTKTFEQVNTIEVSSSHVIKAMWHPKLNQIFVGCGNGVVKCYYDKKRSLRGAKLCIIKTHRKKDTIEVVSTQQIITPHALPLFRQEKLRTSKKKMEKERLDPVKSHRPDLPITSGQGGRVAASGSTLSSFVIRNLGLSKRVNDEQDPREAILKYAKDAEENPFWVAPAYKKTQPKPIFQGSDEEGPSDPKKSKTS
Summary
Similarity
Belongs to the WD repeat GAD-1 family.
Keywords
Complete proteome
Reference proteome
Repeat
WD repeat
Feature
chain Gastrulation defective protein 1 homolog
Uniprot
H9JHZ6
A0A2H1W2P4
A0A2W1BY87
A0A194QHY1
A0A212FPX0
A0A0L7LD21
+ More
A0A194QT25 A0A088AHJ2 A0A2A3EC20 A0A2P8ZF38 A0A0L0CT61 A0A0M9A8Z3 A0A0C9QU76 A0A0C9QKW8 W8BIK9 A0A034WR59 A0A151WST6 E2A3E8 A0A026W4V6 A0A151IAZ0 A0A195BS28 B3NPS5 A0A151J2K2 A0A195EW76 A0A158NNS2 A0A1W4UMK4 A0A154PS53 F4WV22 A0A0A1XM26 B4QAZ6 Q9W1J3 A0A0M5IXN1 B4PA68 A0A1J1HLA3 B4I8T4 A0A1B0G7Q0 A0A1A9ZWD7 B4LM82 B4KNM9 A0A1B0BNI4 A0A232EYR8 A0A1A9XP27 A0A1A9V8D3 A0A2J7RSL7 A0A3B0JRH9 Q17KF1 K7JAQ6 A0A1I8M6J9 B4J6L5 B3ME80 A0A182HC20 B4MYD5 A0A1A9WIQ5 A0A1I8PI69 A0A182F154 A0A0L7RID2 Q293F7 B4GBL0 A0A182GHW5 W5JU00 A0A2M4BH90 A0A067R629 A0A0K8VWI8 A0A1Q3F4V0 A0A1Q3F4R8 A0A182W6T6 A0A182QZ65 B0XJC0 A0A182K3M9 A0A182L130 A0A182J5K2 A0A182PCL1 A0A182V9Q6 A0A182XL15 A0A182Y3K2 A0A182R556 A0A336MEL8 A0A182UBW8 A0A336ML75 E9J9M8 A0A182HUE0 A0A182MYW5 A0A084W4P4 A0A1Y1LFM9 A0A2M4AKS9 A0A2M4AJS2 A0A1W4XFF2 E0VYP6 A0A1W4XEI4 A0A182M5F5 D7EJE9 A0A1B6H134 A0A1B6IFT2 A0A1B6HCS6 A0A1B6M4Z0 A0A087U8Q1 A0A1B6DWZ1 A0A0A9W9H9 A0A0K8SN21
A0A194QT25 A0A088AHJ2 A0A2A3EC20 A0A2P8ZF38 A0A0L0CT61 A0A0M9A8Z3 A0A0C9QU76 A0A0C9QKW8 W8BIK9 A0A034WR59 A0A151WST6 E2A3E8 A0A026W4V6 A0A151IAZ0 A0A195BS28 B3NPS5 A0A151J2K2 A0A195EW76 A0A158NNS2 A0A1W4UMK4 A0A154PS53 F4WV22 A0A0A1XM26 B4QAZ6 Q9W1J3 A0A0M5IXN1 B4PA68 A0A1J1HLA3 B4I8T4 A0A1B0G7Q0 A0A1A9ZWD7 B4LM82 B4KNM9 A0A1B0BNI4 A0A232EYR8 A0A1A9XP27 A0A1A9V8D3 A0A2J7RSL7 A0A3B0JRH9 Q17KF1 K7JAQ6 A0A1I8M6J9 B4J6L5 B3ME80 A0A182HC20 B4MYD5 A0A1A9WIQ5 A0A1I8PI69 A0A182F154 A0A0L7RID2 Q293F7 B4GBL0 A0A182GHW5 W5JU00 A0A2M4BH90 A0A067R629 A0A0K8VWI8 A0A1Q3F4V0 A0A1Q3F4R8 A0A182W6T6 A0A182QZ65 B0XJC0 A0A182K3M9 A0A182L130 A0A182J5K2 A0A182PCL1 A0A182V9Q6 A0A182XL15 A0A182Y3K2 A0A182R556 A0A336MEL8 A0A182UBW8 A0A336ML75 E9J9M8 A0A182HUE0 A0A182MYW5 A0A084W4P4 A0A1Y1LFM9 A0A2M4AKS9 A0A2M4AJS2 A0A1W4XFF2 E0VYP6 A0A1W4XEI4 A0A182M5F5 D7EJE9 A0A1B6H134 A0A1B6IFT2 A0A1B6HCS6 A0A1B6M4Z0 A0A087U8Q1 A0A1B6DWZ1 A0A0A9W9H9 A0A0K8SN21
Pubmed
19121390
28756777
26354079
22118469
26227816
29403074
+ More
26108605 24495485 25348373 20798317 24508170 30249741 17994087 21347285 21719571 25830018 22936249 10731132 12537572 12537569 17550304 28648823 17510324 20075255 25315136 26483478 15632085 20920257 23761445 24845553 20966253 25244985 21282665 24438588 28004739 20566863 18362917 19820115 25401762 26823975
26108605 24495485 25348373 20798317 24508170 30249741 17994087 21347285 21719571 25830018 22936249 10731132 12537572 12537569 17550304 28648823 17510324 20075255 25315136 26483478 15632085 20920257 23761445 24845553 20966253 25244985 21282665 24438588 28004739 20566863 18362917 19820115 25401762 26823975
EMBL
BABH01035297
ODYU01005877
SOQ47216.1
KZ149947
PZC76733.1
KQ459193
+ More
KPJ03061.1 AGBW02000859 OWR55753.1 JTDY01001610 KOB73367.1 KQ461154 KPJ08622.1 KZ288310 PBC28591.1 PYGN01000076 PSN55116.1 JRES01000049 KNC34604.1 KQ435711 KOX79660.1 GBYB01007264 JAG77031.1 GBYB01001207 JAG70974.1 GAMC01013444 JAB93111.1 GAKP01002125 JAC56827.1 KQ982766 KYQ50982.1 GL436428 EFN72012.1 KK107485 QOIP01000006 EZA50084.1 RLU21933.1 KQ978169 KYM96608.1 KQ976417 KYM89820.1 CH954179 EDV56866.1 KQ980382 KYN16309.1 KQ981953 KYN32488.1 ADTU01021742 KQ435119 KZC14731.1 GL888384 EGI61910.1 GBXI01002280 JAD12012.1 CM000362 CM002911 EDX08433.1 KMY96119.1 AE013599 AY058624 AY113366 CP012524 ALC41798.1 CM000158 EDW92394.1 CVRI01000010 CRK88715.1 CH480824 EDW57009.1 CCAG010016890 CH940648 EDW60960.1 CH933808 EDW10014.1 JXJN01017413 NNAY01001609 OXU23440.1 NEVH01000253 PNF43828.1 OUUW01000001 SPP75946.1 CH477225 EAT47139.1 AAZX01000978 CH916367 EDW00918.1 CH902619 EDV35475.1 JXUM01033196 KQ560981 KXJ80146.1 CH963894 EDW77124.1 KQ414583 KOC70727.1 CM000071 EAL24654.2 CH479181 EDW31305.1 JXUM01012233 KQ560349 KXJ82902.1 ADMH02000146 ETN67611.1 GGFJ01003027 MBW52168.1 KK852936 KDR13599.1 GDHF01009090 JAI43224.1 GFDL01012459 JAV22586.1 GFDL01012518 JAV22527.1 AXCN02000817 DS233478 EDS30157.1 UFQT01000450 SSX24508.1 UFQS01001558 UFQT01001558 SSX11439.1 SSX31006.1 GL769399 EFZ10467.1 APCN01002453 ATLV01020363 ATLV01020364 KE525299 KFB45188.1 GEZM01058747 JAV71691.1 GGFK01008011 MBW41332.1 GGFK01007719 MBW41040.1 DS235845 EEB18502.1 AXCM01012135 DS497704 EFA12723.1 GECZ01001408 JAS68361.1 GECU01021930 JAS85776.1 GECU01035223 JAS72483.1 GEBQ01008981 JAT30996.1 KK118740 KFM73740.1 GEDC01007121 JAS30177.1 GBHO01040471 JAG03133.1 GBRD01011108 GDHC01008383 JAG54716.1 JAQ10246.1
KPJ03061.1 AGBW02000859 OWR55753.1 JTDY01001610 KOB73367.1 KQ461154 KPJ08622.1 KZ288310 PBC28591.1 PYGN01000076 PSN55116.1 JRES01000049 KNC34604.1 KQ435711 KOX79660.1 GBYB01007264 JAG77031.1 GBYB01001207 JAG70974.1 GAMC01013444 JAB93111.1 GAKP01002125 JAC56827.1 KQ982766 KYQ50982.1 GL436428 EFN72012.1 KK107485 QOIP01000006 EZA50084.1 RLU21933.1 KQ978169 KYM96608.1 KQ976417 KYM89820.1 CH954179 EDV56866.1 KQ980382 KYN16309.1 KQ981953 KYN32488.1 ADTU01021742 KQ435119 KZC14731.1 GL888384 EGI61910.1 GBXI01002280 JAD12012.1 CM000362 CM002911 EDX08433.1 KMY96119.1 AE013599 AY058624 AY113366 CP012524 ALC41798.1 CM000158 EDW92394.1 CVRI01000010 CRK88715.1 CH480824 EDW57009.1 CCAG010016890 CH940648 EDW60960.1 CH933808 EDW10014.1 JXJN01017413 NNAY01001609 OXU23440.1 NEVH01000253 PNF43828.1 OUUW01000001 SPP75946.1 CH477225 EAT47139.1 AAZX01000978 CH916367 EDW00918.1 CH902619 EDV35475.1 JXUM01033196 KQ560981 KXJ80146.1 CH963894 EDW77124.1 KQ414583 KOC70727.1 CM000071 EAL24654.2 CH479181 EDW31305.1 JXUM01012233 KQ560349 KXJ82902.1 ADMH02000146 ETN67611.1 GGFJ01003027 MBW52168.1 KK852936 KDR13599.1 GDHF01009090 JAI43224.1 GFDL01012459 JAV22586.1 GFDL01012518 JAV22527.1 AXCN02000817 DS233478 EDS30157.1 UFQT01000450 SSX24508.1 UFQS01001558 UFQT01001558 SSX11439.1 SSX31006.1 GL769399 EFZ10467.1 APCN01002453 ATLV01020363 ATLV01020364 KE525299 KFB45188.1 GEZM01058747 JAV71691.1 GGFK01008011 MBW41332.1 GGFK01007719 MBW41040.1 DS235845 EEB18502.1 AXCM01012135 DS497704 EFA12723.1 GECZ01001408 JAS68361.1 GECU01021930 JAS85776.1 GECU01035223 JAS72483.1 GEBQ01008981 JAT30996.1 KK118740 KFM73740.1 GEDC01007121 JAS30177.1 GBHO01040471 JAG03133.1 GBRD01011108 GDHC01008383 JAG54716.1 JAQ10246.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000037510
UP000053240
UP000005203
+ More
UP000242457 UP000245037 UP000037069 UP000053105 UP000075809 UP000000311 UP000053097 UP000279307 UP000078542 UP000078540 UP000008711 UP000078492 UP000078541 UP000005205 UP000192221 UP000076502 UP000007755 UP000000304 UP000000803 UP000092553 UP000002282 UP000183832 UP000001292 UP000092444 UP000092445 UP000008792 UP000009192 UP000092460 UP000215335 UP000092443 UP000078200 UP000235965 UP000268350 UP000008820 UP000002358 UP000095301 UP000001070 UP000007801 UP000069940 UP000249989 UP000007798 UP000091820 UP000095300 UP000069272 UP000053825 UP000001819 UP000008744 UP000000673 UP000027135 UP000075920 UP000075886 UP000002320 UP000075881 UP000075882 UP000075880 UP000075885 UP000075903 UP000076407 UP000076408 UP000075900 UP000075902 UP000075840 UP000075884 UP000030765 UP000192223 UP000009046 UP000075883 UP000007266 UP000054359
UP000242457 UP000245037 UP000037069 UP000053105 UP000075809 UP000000311 UP000053097 UP000279307 UP000078542 UP000078540 UP000008711 UP000078492 UP000078541 UP000005205 UP000192221 UP000076502 UP000007755 UP000000304 UP000000803 UP000092553 UP000002282 UP000183832 UP000001292 UP000092444 UP000092445 UP000008792 UP000009192 UP000092460 UP000215335 UP000092443 UP000078200 UP000235965 UP000268350 UP000008820 UP000002358 UP000095301 UP000001070 UP000007801 UP000069940 UP000249989 UP000007798 UP000091820 UP000095300 UP000069272 UP000053825 UP000001819 UP000008744 UP000000673 UP000027135 UP000075920 UP000075886 UP000002320 UP000075881 UP000075882 UP000075880 UP000075885 UP000075903 UP000076407 UP000076408 UP000075900 UP000075902 UP000075840 UP000075884 UP000030765 UP000192223 UP000009046 UP000075883 UP000007266 UP000054359
Interpro
IPR001680
WD40_repeat
+ More
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR020472 G-protein_beta_WD-40_rep
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR026906 LRR_5
IPR032675 LRR_dom_sf
IPR033140 Lipase_GDXG_put_SER_AS
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR020472 G-protein_beta_WD-40_rep
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR026906 LRR_5
IPR032675 LRR_dom_sf
IPR033140 Lipase_GDXG_put_SER_AS
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
H9JHZ6
A0A2H1W2P4
A0A2W1BY87
A0A194QHY1
A0A212FPX0
A0A0L7LD21
+ More
A0A194QT25 A0A088AHJ2 A0A2A3EC20 A0A2P8ZF38 A0A0L0CT61 A0A0M9A8Z3 A0A0C9QU76 A0A0C9QKW8 W8BIK9 A0A034WR59 A0A151WST6 E2A3E8 A0A026W4V6 A0A151IAZ0 A0A195BS28 B3NPS5 A0A151J2K2 A0A195EW76 A0A158NNS2 A0A1W4UMK4 A0A154PS53 F4WV22 A0A0A1XM26 B4QAZ6 Q9W1J3 A0A0M5IXN1 B4PA68 A0A1J1HLA3 B4I8T4 A0A1B0G7Q0 A0A1A9ZWD7 B4LM82 B4KNM9 A0A1B0BNI4 A0A232EYR8 A0A1A9XP27 A0A1A9V8D3 A0A2J7RSL7 A0A3B0JRH9 Q17KF1 K7JAQ6 A0A1I8M6J9 B4J6L5 B3ME80 A0A182HC20 B4MYD5 A0A1A9WIQ5 A0A1I8PI69 A0A182F154 A0A0L7RID2 Q293F7 B4GBL0 A0A182GHW5 W5JU00 A0A2M4BH90 A0A067R629 A0A0K8VWI8 A0A1Q3F4V0 A0A1Q3F4R8 A0A182W6T6 A0A182QZ65 B0XJC0 A0A182K3M9 A0A182L130 A0A182J5K2 A0A182PCL1 A0A182V9Q6 A0A182XL15 A0A182Y3K2 A0A182R556 A0A336MEL8 A0A182UBW8 A0A336ML75 E9J9M8 A0A182HUE0 A0A182MYW5 A0A084W4P4 A0A1Y1LFM9 A0A2M4AKS9 A0A2M4AJS2 A0A1W4XFF2 E0VYP6 A0A1W4XEI4 A0A182M5F5 D7EJE9 A0A1B6H134 A0A1B6IFT2 A0A1B6HCS6 A0A1B6M4Z0 A0A087U8Q1 A0A1B6DWZ1 A0A0A9W9H9 A0A0K8SN21
A0A194QT25 A0A088AHJ2 A0A2A3EC20 A0A2P8ZF38 A0A0L0CT61 A0A0M9A8Z3 A0A0C9QU76 A0A0C9QKW8 W8BIK9 A0A034WR59 A0A151WST6 E2A3E8 A0A026W4V6 A0A151IAZ0 A0A195BS28 B3NPS5 A0A151J2K2 A0A195EW76 A0A158NNS2 A0A1W4UMK4 A0A154PS53 F4WV22 A0A0A1XM26 B4QAZ6 Q9W1J3 A0A0M5IXN1 B4PA68 A0A1J1HLA3 B4I8T4 A0A1B0G7Q0 A0A1A9ZWD7 B4LM82 B4KNM9 A0A1B0BNI4 A0A232EYR8 A0A1A9XP27 A0A1A9V8D3 A0A2J7RSL7 A0A3B0JRH9 Q17KF1 K7JAQ6 A0A1I8M6J9 B4J6L5 B3ME80 A0A182HC20 B4MYD5 A0A1A9WIQ5 A0A1I8PI69 A0A182F154 A0A0L7RID2 Q293F7 B4GBL0 A0A182GHW5 W5JU00 A0A2M4BH90 A0A067R629 A0A0K8VWI8 A0A1Q3F4V0 A0A1Q3F4R8 A0A182W6T6 A0A182QZ65 B0XJC0 A0A182K3M9 A0A182L130 A0A182J5K2 A0A182PCL1 A0A182V9Q6 A0A182XL15 A0A182Y3K2 A0A182R556 A0A336MEL8 A0A182UBW8 A0A336ML75 E9J9M8 A0A182HUE0 A0A182MYW5 A0A084W4P4 A0A1Y1LFM9 A0A2M4AKS9 A0A2M4AJS2 A0A1W4XFF2 E0VYP6 A0A1W4XEI4 A0A182M5F5 D7EJE9 A0A1B6H134 A0A1B6IFT2 A0A1B6HCS6 A0A1B6M4Z0 A0A087U8Q1 A0A1B6DWZ1 A0A0A9W9H9 A0A0K8SN21
PDB
6QDV
E-value=2.06202e-12,
Score=177
Ontologies
GO
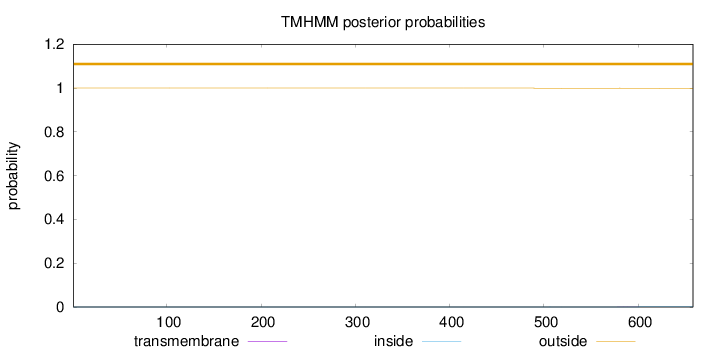
Topology
Length:
658
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00886
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00031
outside
1 - 658
Population Genetic Test Statistics
Pi
172.907138
Theta
160.02021
Tajima's D
1.036144
CLR
2.539625
CSRT
0.668766561671916
Interpretation
Uncertain
