Gene
KWMTBOMO12258 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009139
Annotation
Counting_factor_associated_protein_D_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 1.727 Lysosomal Reliability : 1.368
Sequence
CDS
ATGTTATTCGCTGCGTTGTTGTGTTTTTTCGTGGCCTTCGCATCTGGATATGTCGATAAAGGGTCGCCACCACAATGGAGTCCAGTATACACAGTAAAGGGACTCCTAAATATACCCTATGCTGAACTGCATGAGCCATTCTATGCTTGGTACGACAGTAAGAACAGCAAATCAAGAATCGATTACTATGGAGGTATGGTCAAGACTTACCAGTTCACGTCAGCGGTCTACCCACCCTACGGTACTTCTATAAAGATCGCTCCGGTCACAACGGAAACCGAGATGAACAAGGAGACGTGCCTGCAAGTCAATTCGACGCAAGATCAGCTCCAAGACATCCAGTCAGTGCTACCGGACATGACCGATTTTAAATATATCGGCACAGAAACAATGCAGGATGCCGACACGGCCAAATGGCAGATGGTACAGCCGGTCGGCGATAAGCTAAACAAATACACGATGTGGGTTAAATACAAGAAGACTTTGAAAGGGGATTCTGTGCCCATACCGGTCCGATATGAGATGAAAGGTTTCAATTCGCTCCTGGGCTCCCACTACGATCATTACTATCTGGATTATGAAGATTACAATATTGATGAGATCGATCCTGACGTCTTCAAAGTTGACAGTAACATGCAGTGCACTGGGTTCCCGGGGCCCGGCTCGCGACACTTCGCCACCTTCAACCCGATGAAGGAGTTCGTGCGGCCCGTGCACGACGCCCACGTCCACGACGAGTTCGAGCGTTTCAAAGTCAAACACCAGAGGCAGTACGCGAGCGACCTGGAGCACGAGAAGAGGCTGAACATCTTCAGGCAGTCGCTCAGATACATACATTCCAATAATCGAGCGAACCGCGGTTTCACCATGTCCGTGAACCATCTTGCCGATCGCACTGACGACGAGCTCGCCGCCCTCAGAGGGAGGAGGTACTCGGGGCCCAGCCCTCACGGTCTCCCGTTCCCGTACAGCAAATCTCGAGTGGAGGAGTTGAGCGTAAAGCTGCCTCCGGAACACGACTGGAGATTGTTCGGAGCGGTCACTCCCGTTAAAGATCAGTCGGTGTGCGGCTCGTGCTGGTCGTTCGGCACGGTGGGCGCGGTGGAGGGCGCGCTGTTCCTGCACAACGGCGGACACCTGGTCCGGCTCAGCCAGCAGGCGCTCATCGACTGCTCCTGGGGTTTCGGGAACAACGGCTGCGACGGAGGCGAGGACTTCAGGGCCTACGAGTGGATTAAGAGGCACGGCCTCCCCACAGAGGAAGACTACGGAGGATACCTCGGACAGGATGGCTACTGTCATGTCGATAACGTGACGGCTGTCACCTCGATCACCGGCTGGGTTAACGTGACGACAAACGATGAGAAGGCGCTAAAGTTGGCGATCTTCAAGCACGGTCCGATTTCGGTGGCCATCGACGCTTCCCACAAGAGCTTCAGCTTCTACGCGAACGGAGTGTATTTTGAACCGAATTGCAAGAACAAAGTTGAGGAGCTGGACCACGCGGTGCTGGCGGTCGGCTACGGCGTCCTCAACGGCAAGAAGTACTGGCTCATCAAGAACTCCTGGTCGAACATGTGGGGGAACGACGGCTACGTCCTCATGTCCTCGAAGGAGAACAACTGCGGAGTCGAAAGTGCACCCACATACACCACGTGCATGCTGAAGATCAGTAAATTGTCGCAGCGGCGATTCCTCATGCGATAA
Protein
MLFAALLCFFVAFASGYVDKGSPPQWSPVYTVKGLLNIPYAELHEPFYAWYDSKNSKSRIDYYGGMVKTYQFTSAVYPPYGTSIKIAPVTTETEMNKETCLQVNSTQDQLQDIQSVLPDMTDFKYIGTETMQDADTAKWQMVQPVGDKLNKYTMWVKYKKTLKGDSVPIPVRYEMKGFNSLLGSHYDHYYLDYEDYNIDEIDPDVFKVDSNMQCTGFPGPGSRHFATFNPMKEFVRPVHDAHVHDEFERFKVKHQRQYASDLEHEKRLNIFRQSLRYIHSNNRANRGFTMSVNHLADRTDDELAALRGRRYSGPSPHGLPFPYSKSRVEELSVKLPPEHDWRLFGAVTPVKDQSVCGSCWSFGTVGAVEGALFLHNGGHLVRLSQQALIDCSWGFGNNGCDGGEDFRAYEWIKRHGLPTEEDYGGYLGQDGYCHVDNVTAVTSITGWVNVTTNDEKALKLAIFKHGPISVAIDASHKSFSFYANGVYFEPNCKNKVEELDHAVLAVGYGVLNGKKYWLIKNSWSNMWGNDGYVLMSSKENNCGVESAPTYTTCMLKISKLSQRRFLMR
Summary
Similarity
Belongs to the peptidase C1 family.
Belongs to the tubulin family.
Belongs to the tubulin family.
Uniprot
H9JHZ1
A0A194QC71
S4PTE3
A0A3S2LHA4
A0A2W1BNV9
A0A2H1W2A2
+ More
A0A212FPU5 D6WRI7 A0A0B5IZ56 R4G406 A0A171AH64 V5GU72 A0A1Y1NIN4 A0A224XHL3 J3JWF1 Q9U914 A0A182YA94 A0A1L8DPF8 A0A182KUV8 A0A182K2U4 A0A182WRD1 Q7QL68 A0A182VKN0 A0A0C9RUS1 A0A034VUB0 A0A182M8H3 B3MAG6 A0A182MY79 B4MLV9 A0A182PBW2 A0A1L8EEE7 A0A182WEA7 A0A0K8TWU9 B4L168 A0A182QBZ1 A0A182J0S4 A0A067QZ64 T1P8Q0 A0A0L7QVA4 A0A182RLX7 A0A0L0BV98 A0A2M4BJK5 A0A2M4BJI3 B4QJI8 Q9V3U6 T1DG71 A0A1I8NES7 B4PHY4 A0A084W8F1 B4HHA4 A0A2M4AIZ6 A0A1W4U6M1 A0A1I8NLT1 O97453 B3NCZ7 B4LC35 A0A2J7QI59 A0A182FW06 A0A2M3Z3H1 A0A2M3ZE63 A0A182I650 N6UA25 A0A0A1XPY6 A0A3B0JSY6 A0A2M4DNC7 A0A2M4DPK4 A0A0M3QW40 U4UHC6 B0XCU8 U5EWS0 A0A232EZY9 K7J3M0 A0A1Q3F4S5 A0A1J1J120 B4IXZ3 Q2M011 A0A1A9WE80 A0A3L8E0N5 A0A1A9Y5W1 A0A1A9US28 A0A1B0BAT2 A0A154NY45 A0A1B0GD03 Q176E8 A0A1S4FDC8 A0A151IC91 A0A1B0AJE0 A0A182H6B8 A0A182HAH0 A0A0J7L7V5 Q1PA54 A0A151WRQ3 Q2PZ08 A0A151JVY2 T1HS97 A0A1W4X8X7 A0A195BNR5 A0A158N9R4 E2AE51 E9J0U7
A0A212FPU5 D6WRI7 A0A0B5IZ56 R4G406 A0A171AH64 V5GU72 A0A1Y1NIN4 A0A224XHL3 J3JWF1 Q9U914 A0A182YA94 A0A1L8DPF8 A0A182KUV8 A0A182K2U4 A0A182WRD1 Q7QL68 A0A182VKN0 A0A0C9RUS1 A0A034VUB0 A0A182M8H3 B3MAG6 A0A182MY79 B4MLV9 A0A182PBW2 A0A1L8EEE7 A0A182WEA7 A0A0K8TWU9 B4L168 A0A182QBZ1 A0A182J0S4 A0A067QZ64 T1P8Q0 A0A0L7QVA4 A0A182RLX7 A0A0L0BV98 A0A2M4BJK5 A0A2M4BJI3 B4QJI8 Q9V3U6 T1DG71 A0A1I8NES7 B4PHY4 A0A084W8F1 B4HHA4 A0A2M4AIZ6 A0A1W4U6M1 A0A1I8NLT1 O97453 B3NCZ7 B4LC35 A0A2J7QI59 A0A182FW06 A0A2M3Z3H1 A0A2M3ZE63 A0A182I650 N6UA25 A0A0A1XPY6 A0A3B0JSY6 A0A2M4DNC7 A0A2M4DPK4 A0A0M3QW40 U4UHC6 B0XCU8 U5EWS0 A0A232EZY9 K7J3M0 A0A1Q3F4S5 A0A1J1J120 B4IXZ3 Q2M011 A0A1A9WE80 A0A3L8E0N5 A0A1A9Y5W1 A0A1A9US28 A0A1B0BAT2 A0A154NY45 A0A1B0GD03 Q176E8 A0A1S4FDC8 A0A151IC91 A0A1B0AJE0 A0A182H6B8 A0A182HAH0 A0A0J7L7V5 Q1PA54 A0A151WRQ3 Q2PZ08 A0A151JVY2 T1HS97 A0A1W4X8X7 A0A195BNR5 A0A158N9R4 E2AE51 E9J0U7
Pubmed
19121390
26354079
23622113
28756777
22118469
18362917
+ More
19820115 28004739 22516182 10050046 25244985 20966253 9087549 12364791 14747013 17210077 25348373 17994087 24845553 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17550304 24438588 23537049 25830018 28648823 20075255 15632085 30249741 17510324 26483478 16907828 20353571 21347285 20798317 21282665
19820115 28004739 22516182 10050046 25244985 20966253 9087549 12364791 14747013 17210077 25348373 17994087 24845553 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17550304 24438588 23537049 25830018 28648823 20075255 15632085 30249741 17510324 26483478 16907828 20353571 21347285 20798317 21282665
EMBL
BABH01035287
BABH01035288
KQ459193
KPJ03062.1
GAIX01012188
JAA80372.1
+ More
RSAL01000122 RVE46658.1 KZ149947 PZC76732.1 ODYU01005877 SOQ47215.1 AGBW02000859 OWR55754.1 KQ971351 EFA07493.2 KP303285 AJF94883.1 GAHY01001760 JAA75750.1 GEMB01001072 JAS02075.1 GALX01004673 JAB63793.1 GEZM01003865 JAV96515.1 GFTR01007128 JAW09298.1 BT127569 AEE62531.1 AB011377 BAA86911.1 GFDF01005803 JAV08281.1 AAAB01007910 EAA03137.4 GBYB01011386 JAG81153.1 KU904503 GAKP01012036 ANK58172.1 JAC46916.1 AXCM01001764 CH902618 EDV40217.1 CH963847 EDW73170.1 GFDG01001785 JAV17014.1 GDHF01033576 GDHF01008004 JAI18738.1 JAI44310.1 CH933809 EDW19250.1 AXCN02002037 KK853083 KDR11614.1 KA644495 AFP59124.1 KQ414731 KOC62391.1 JRES01001278 KNC23990.1 GGFJ01004022 MBW53163.1 GGFJ01004023 MBW53164.1 CM000363 CM002912 EDX10397.1 KMY99497.1 AE014296 AY122222 AB011376 AAF49777.1 AAM52734.1 BAA86910.1 GAMD01002876 JAA98714.1 CM000159 EDW94459.1 ATLV01021421 KE525318 KFB46495.1 CH480815 EDW41429.1 GGFK01007396 MBW40717.1 AB011375 BAA76272.1 CH954178 EDV51719.1 CH940647 EDW70863.1 NEVH01013960 PNF28261.1 GGFM01002315 MBW23066.1 GGFM01005969 MBW26720.1 APCN01003562 APGK01036954 APGK01036955 KB740945 ENN77491.1 GBXI01008134 GBXI01000913 JAD06158.1 JAD13379.1 OUUW01000002 SPP76476.1 GGFL01014861 MBW79039.1 GGFL01014860 MBW79038.1 CP012525 ALC43483.1 KB632197 ERL90006.1 DS232720 EDS45115.1 GANO01000430 JAB59441.1 NNAY01001475 OXU23840.1 GFDL01012479 JAV22566.1 CVRI01000066 CRL06227.1 CH916366 EDV97536.1 CH379069 EAL31126.1 QOIP01000001 RLU26281.1 JXJN01011148 KQ434782 KZC04596.1 CCAG010002253 CCAG010002254 CCAG010002255 CH477388 EAT42028.1 KQ978067 KYM97671.1 JXUM01113753 KQ565722 KXJ70744.1 JXUM01122736 KQ566637 KXJ69956.1 LBMM01000340 KMR00225.1 DQ459305 ABE72972.1 KQ982796 KYQ50589.1 DQ294221 EZ422242 ABC48937.1 ADD18518.1 KQ981672 KYN38426.1 ACPB03005923 KQ976428 KYM87862.1 ADTU01009804 ADTU01009805 GL438827 EFN68284.1 GL767538 EFZ13575.1
RSAL01000122 RVE46658.1 KZ149947 PZC76732.1 ODYU01005877 SOQ47215.1 AGBW02000859 OWR55754.1 KQ971351 EFA07493.2 KP303285 AJF94883.1 GAHY01001760 JAA75750.1 GEMB01001072 JAS02075.1 GALX01004673 JAB63793.1 GEZM01003865 JAV96515.1 GFTR01007128 JAW09298.1 BT127569 AEE62531.1 AB011377 BAA86911.1 GFDF01005803 JAV08281.1 AAAB01007910 EAA03137.4 GBYB01011386 JAG81153.1 KU904503 GAKP01012036 ANK58172.1 JAC46916.1 AXCM01001764 CH902618 EDV40217.1 CH963847 EDW73170.1 GFDG01001785 JAV17014.1 GDHF01033576 GDHF01008004 JAI18738.1 JAI44310.1 CH933809 EDW19250.1 AXCN02002037 KK853083 KDR11614.1 KA644495 AFP59124.1 KQ414731 KOC62391.1 JRES01001278 KNC23990.1 GGFJ01004022 MBW53163.1 GGFJ01004023 MBW53164.1 CM000363 CM002912 EDX10397.1 KMY99497.1 AE014296 AY122222 AB011376 AAF49777.1 AAM52734.1 BAA86910.1 GAMD01002876 JAA98714.1 CM000159 EDW94459.1 ATLV01021421 KE525318 KFB46495.1 CH480815 EDW41429.1 GGFK01007396 MBW40717.1 AB011375 BAA76272.1 CH954178 EDV51719.1 CH940647 EDW70863.1 NEVH01013960 PNF28261.1 GGFM01002315 MBW23066.1 GGFM01005969 MBW26720.1 APCN01003562 APGK01036954 APGK01036955 KB740945 ENN77491.1 GBXI01008134 GBXI01000913 JAD06158.1 JAD13379.1 OUUW01000002 SPP76476.1 GGFL01014861 MBW79039.1 GGFL01014860 MBW79038.1 CP012525 ALC43483.1 KB632197 ERL90006.1 DS232720 EDS45115.1 GANO01000430 JAB59441.1 NNAY01001475 OXU23840.1 GFDL01012479 JAV22566.1 CVRI01000066 CRL06227.1 CH916366 EDV97536.1 CH379069 EAL31126.1 QOIP01000001 RLU26281.1 JXJN01011148 KQ434782 KZC04596.1 CCAG010002253 CCAG010002254 CCAG010002255 CH477388 EAT42028.1 KQ978067 KYM97671.1 JXUM01113753 KQ565722 KXJ70744.1 JXUM01122736 KQ566637 KXJ69956.1 LBMM01000340 KMR00225.1 DQ459305 ABE72972.1 KQ982796 KYQ50589.1 DQ294221 EZ422242 ABC48937.1 ADD18518.1 KQ981672 KYN38426.1 ACPB03005923 KQ976428 KYM87862.1 ADTU01009804 ADTU01009805 GL438827 EFN68284.1 GL767538 EFZ13575.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000007151
UP000007266
UP000076408
+ More
UP000075882 UP000075881 UP000076407 UP000007062 UP000075903 UP000075883 UP000007801 UP000075884 UP000007798 UP000075885 UP000075920 UP000009192 UP000075886 UP000075880 UP000027135 UP000053825 UP000075900 UP000037069 UP000000304 UP000000803 UP000095301 UP000002282 UP000030765 UP000001292 UP000192221 UP000095300 UP000008711 UP000008792 UP000235965 UP000069272 UP000075840 UP000019118 UP000268350 UP000092553 UP000030742 UP000002320 UP000215335 UP000002358 UP000183832 UP000001070 UP000001819 UP000091820 UP000279307 UP000092443 UP000078200 UP000092460 UP000076502 UP000092444 UP000008820 UP000078542 UP000092445 UP000069940 UP000249989 UP000036403 UP000075809 UP000078541 UP000015103 UP000192223 UP000078540 UP000005205 UP000000311
UP000075882 UP000075881 UP000076407 UP000007062 UP000075903 UP000075883 UP000007801 UP000075884 UP000007798 UP000075885 UP000075920 UP000009192 UP000075886 UP000075880 UP000027135 UP000053825 UP000075900 UP000037069 UP000000304 UP000000803 UP000095301 UP000002282 UP000030765 UP000001292 UP000192221 UP000095300 UP000008711 UP000008792 UP000235965 UP000069272 UP000075840 UP000019118 UP000268350 UP000092553 UP000030742 UP000002320 UP000215335 UP000002358 UP000183832 UP000001070 UP000001819 UP000091820 UP000279307 UP000092443 UP000078200 UP000092460 UP000076502 UP000092444 UP000008820 UP000078542 UP000092445 UP000069940 UP000249989 UP000036403 UP000075809 UP000078541 UP000015103 UP000192223 UP000078540 UP000005205 UP000000311
PRIDE
Pfam
Interpro
IPR025661
Pept_asp_AS
+ More
IPR025660 Pept_his_AS
IPR000668 Peptidase_C1A_C
IPR000169 Pept_cys_AS
IPR038765 Papain-like_cys_pep_sf
IPR013201 Prot_inhib_I29
IPR039417 Peptidase_C1A_papain-like
IPR031305 Casein_CS
IPR023393 START-like_dom_sf
IPR011993 PH-like_dom_sf
IPR001849 PH_domain
IPR002913 START_lipid-bd_dom
IPR036525 Tubulin/FtsZ_GTPase_sf
IPR000217 Tubulin
IPR018316 Tubulin/FtsZ_2-layer-sand-dom
IPR023123 Tubulin_C
IPR002453 Beta_tubulin
IPR037103 Tubulin/FtsZ_C_sf
IPR003008 Tubulin_FtsZ_GTPase
IPR008280 Tub_FtsZ_C
IPR029404 DUF_TPD
IPR039016 RECK
IPR002350 Kazal_dom
IPR036058 Kazal_dom_sf
IPR025660 Pept_his_AS
IPR000668 Peptidase_C1A_C
IPR000169 Pept_cys_AS
IPR038765 Papain-like_cys_pep_sf
IPR013201 Prot_inhib_I29
IPR039417 Peptidase_C1A_papain-like
IPR031305 Casein_CS
IPR023393 START-like_dom_sf
IPR011993 PH-like_dom_sf
IPR001849 PH_domain
IPR002913 START_lipid-bd_dom
IPR036525 Tubulin/FtsZ_GTPase_sf
IPR000217 Tubulin
IPR018316 Tubulin/FtsZ_2-layer-sand-dom
IPR023123 Tubulin_C
IPR002453 Beta_tubulin
IPR037103 Tubulin/FtsZ_C_sf
IPR003008 Tubulin_FtsZ_GTPase
IPR008280 Tub_FtsZ_C
IPR029404 DUF_TPD
IPR039016 RECK
IPR002350 Kazal_dom
IPR036058 Kazal_dom_sf
ProteinModelPortal
H9JHZ1
A0A194QC71
S4PTE3
A0A3S2LHA4
A0A2W1BNV9
A0A2H1W2A2
+ More
A0A212FPU5 D6WRI7 A0A0B5IZ56 R4G406 A0A171AH64 V5GU72 A0A1Y1NIN4 A0A224XHL3 J3JWF1 Q9U914 A0A182YA94 A0A1L8DPF8 A0A182KUV8 A0A182K2U4 A0A182WRD1 Q7QL68 A0A182VKN0 A0A0C9RUS1 A0A034VUB0 A0A182M8H3 B3MAG6 A0A182MY79 B4MLV9 A0A182PBW2 A0A1L8EEE7 A0A182WEA7 A0A0K8TWU9 B4L168 A0A182QBZ1 A0A182J0S4 A0A067QZ64 T1P8Q0 A0A0L7QVA4 A0A182RLX7 A0A0L0BV98 A0A2M4BJK5 A0A2M4BJI3 B4QJI8 Q9V3U6 T1DG71 A0A1I8NES7 B4PHY4 A0A084W8F1 B4HHA4 A0A2M4AIZ6 A0A1W4U6M1 A0A1I8NLT1 O97453 B3NCZ7 B4LC35 A0A2J7QI59 A0A182FW06 A0A2M3Z3H1 A0A2M3ZE63 A0A182I650 N6UA25 A0A0A1XPY6 A0A3B0JSY6 A0A2M4DNC7 A0A2M4DPK4 A0A0M3QW40 U4UHC6 B0XCU8 U5EWS0 A0A232EZY9 K7J3M0 A0A1Q3F4S5 A0A1J1J120 B4IXZ3 Q2M011 A0A1A9WE80 A0A3L8E0N5 A0A1A9Y5W1 A0A1A9US28 A0A1B0BAT2 A0A154NY45 A0A1B0GD03 Q176E8 A0A1S4FDC8 A0A151IC91 A0A1B0AJE0 A0A182H6B8 A0A182HAH0 A0A0J7L7V5 Q1PA54 A0A151WRQ3 Q2PZ08 A0A151JVY2 T1HS97 A0A1W4X8X7 A0A195BNR5 A0A158N9R4 E2AE51 E9J0U7
A0A212FPU5 D6WRI7 A0A0B5IZ56 R4G406 A0A171AH64 V5GU72 A0A1Y1NIN4 A0A224XHL3 J3JWF1 Q9U914 A0A182YA94 A0A1L8DPF8 A0A182KUV8 A0A182K2U4 A0A182WRD1 Q7QL68 A0A182VKN0 A0A0C9RUS1 A0A034VUB0 A0A182M8H3 B3MAG6 A0A182MY79 B4MLV9 A0A182PBW2 A0A1L8EEE7 A0A182WEA7 A0A0K8TWU9 B4L168 A0A182QBZ1 A0A182J0S4 A0A067QZ64 T1P8Q0 A0A0L7QVA4 A0A182RLX7 A0A0L0BV98 A0A2M4BJK5 A0A2M4BJI3 B4QJI8 Q9V3U6 T1DG71 A0A1I8NES7 B4PHY4 A0A084W8F1 B4HHA4 A0A2M4AIZ6 A0A1W4U6M1 A0A1I8NLT1 O97453 B3NCZ7 B4LC35 A0A2J7QI59 A0A182FW06 A0A2M3Z3H1 A0A2M3ZE63 A0A182I650 N6UA25 A0A0A1XPY6 A0A3B0JSY6 A0A2M4DNC7 A0A2M4DPK4 A0A0M3QW40 U4UHC6 B0XCU8 U5EWS0 A0A232EZY9 K7J3M0 A0A1Q3F4S5 A0A1J1J120 B4IXZ3 Q2M011 A0A1A9WE80 A0A3L8E0N5 A0A1A9Y5W1 A0A1A9US28 A0A1B0BAT2 A0A154NY45 A0A1B0GD03 Q176E8 A0A1S4FDC8 A0A151IC91 A0A1B0AJE0 A0A182H6B8 A0A182HAH0 A0A0J7L7V5 Q1PA54 A0A151WRQ3 Q2PZ08 A0A151JVY2 T1HS97 A0A1W4X8X7 A0A195BNR5 A0A158N9R4 E2AE51 E9J0U7
PDB
7PCK
E-value=2.4982e-53,
Score=529
Ontologies
GO
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.994225
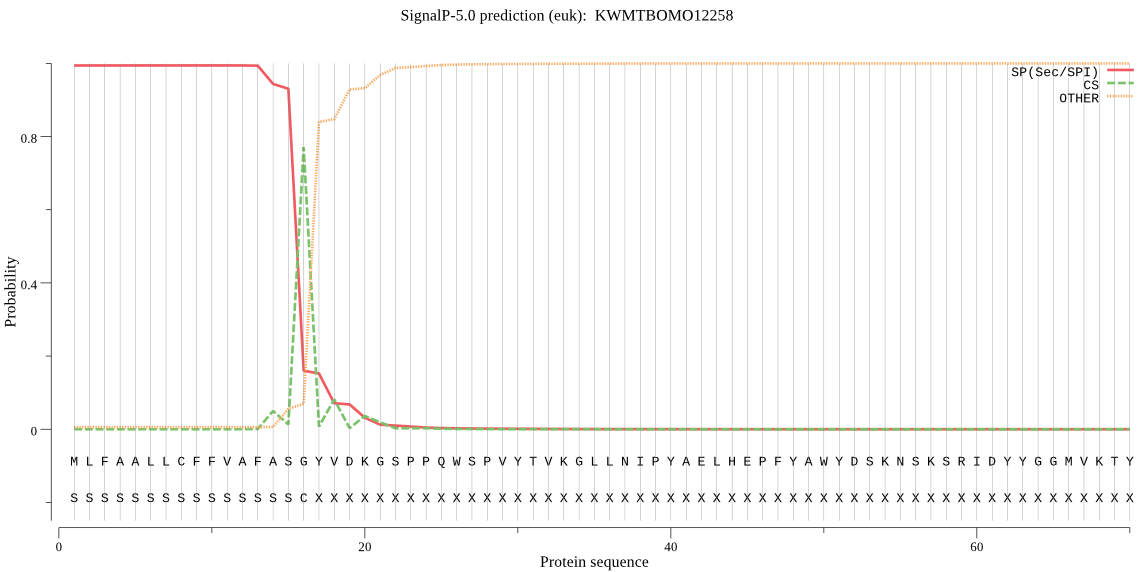
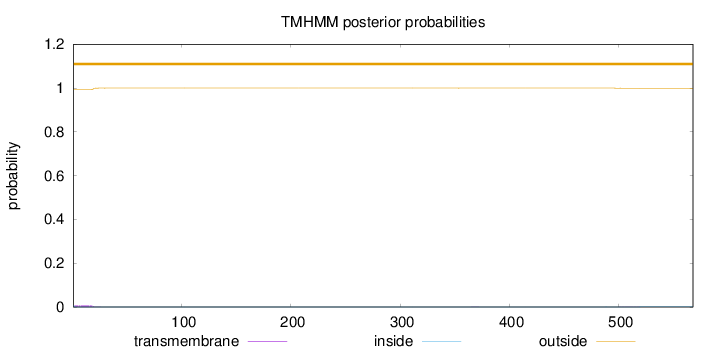
Length:
568
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.141500000000001
Exp number, first 60 AAs:
0.12918
Total prob of N-in:
0.00741
outside
1 - 568
Population Genetic Test Statistics
Pi
188.37185
Theta
176.845668
Tajima's D
1.393175
CLR
0.17578
CSRT
0.76191190440478
Interpretation
Uncertain
