Gene
KWMTBOMO12244
Pre Gene Modal
BGIBMGA014139
Annotation
PREDICTED:_protein_5NUC_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.897
Sequence
CDS
ATGTCTATATTAGCTTTACAAAAGCAGATAAGTGATGCTTCACTCGAAGTGCGTCAAGTCGGAAGGCGTGCAGTGAACCAGCTCGCGGGACGAGGGATGGGCGATGAGGTCATCATCATACACTTCAACGATGTTTACAACATTGAACCGACGACGACCACGGAACCAGTGGGGGGTGCACTGAGATTCAGTACTGCTGTCAAAGCGTTGCAACATTTGAATCCTCTGGTGTTATTCAGCGGGGATGTTTTCTCGCCCAGCATGTTGAGCACGTTCACGAAAGGGGAACAAATGGTGCCTGTATTGAACGAAATAGGAACGCAATGTGCCGTGTTCGGGAACCATGACTTCGATTTCGGTCTAGAAGTCCTGTCGGGCTTAGTAGCCCAATGCAAGTTCCCATGGTTAATGTCCAACGTAATTGACAACGAGACTGGTAGACCTCTTGGTGATGGACGAATAACTCACCCTTACATCGCTAACGGACACAAGATCGGCCTCATAGGTCTCGTGGAACAGGAATGGCTCGAAACCCTGGCCACCATCAACCCCGAAGAGGTGACTTTCATAGACTTCCTCCAGGCTGGATCGAAACTGGCTTCGCAGCTGAAACAAGAGGGCTGTGAGTACGTAATAGCTCTGACACACATGAGAACGCCGAACGACATTAAACTGGCCGAGGGCTGTCCGGACATCGACCTCATACTCGGCGGACACGACCACGTCTACGAGGTCCTTGAGATAAATAACACGTATATAGTAAAAAGCGGCACGGACTTCCGTCAGTTCTCCAAAATCAGCATATCGTTCAAGGGCGAAGCGCCCAAAGTGGAGATAACCGAAGTCCCGGTGACCAGCTCGTATCCCGAAGACGAGGTGCTCAAAGCTAAAGTGGAGAAGTACTCAGCAATGATCGAAGGTAAAATGGACGAGGTATTGGGCAAGTTCAGCGTACCGTTGGAAGGTCGTTTCTCGTACATCCGCCGCATGGAGTGCAACCTCGGCAACTGGGTCACAGACGTGCTGCTGGCGGCCACCGGGGCCGACCTCGTCGTCCTGAACTCGGGAACCTTCCGGTCCGACCAGGTACATCCAGCTGGCGACTTCACTTTACGCGACCTATCCACCATTATACCGATGCGGGATCCGCTCGTGGTGGTGGAAGCCAGCGGCGACGTCATCCTCCAGGTCCTGGAGAACTCTGTCAGCAAATATCCGAGTCTGGAGGGAAGATTCCCTCAGGTTGCCGGTGTGTCATTCGCGTTCGACCCGTCGCTGCCCCCGGGGCGGCGCGTGGCCGAGCACGTGGTGAAGGTCGGCGATGAGTACCTGCAGCGCGCCCAGCGGTACCGCCTCGCCGTCAAGCAGTACCTGCGCGCCGGCAACGACGGCTACACCATGCTGCCCTCCTGCAAGCTACTCGTATCGGAGGACATGTGCCCCGAAATAGGGATGGCGATACAGAACCACTTCGCTGCCATCAACGTGCGCACCGGTAAATCCAAACCTTCGAAGCACAGACAGTCGCTGGTCACGCTCAGCCGGAGGCACAGCTTAGTAAAGATGCTCGATGTAAACGAGCTAGACGGGCCGCCTCCTCTTCGAAGAGCTTCGTCGGCTGCCGTGGAGCCCACTCCACACAGCGGCCATCACAGGCTGACGAGGCGAGCGTCCTTGGACGATCTGGAGCAGCAATCCTGCGAGTTGGCCCCCAAAATAGAGAATAGAATCGTCGTCCTAGATAAACCAGAGAAGCTTCAGGCTTTGCTCGCCGAGCGCGCCCGCTGGGAGTCCGATACAGTCATCAGGGAGGTGGAGGATGAAAACTCACCCTAA
Protein
MSILALQKQISDASLEVRQVGRRAVNQLAGRGMGDEVIIIHFNDVYNIEPTTTTEPVGGALRFSTAVKALQHLNPLVLFSGDVFSPSMLSTFTKGEQMVPVLNEIGTQCAVFGNHDFDFGLEVLSGLVAQCKFPWLMSNVIDNETGRPLGDGRITHPYIANGHKIGLIGLVEQEWLETLATINPEEVTFIDFLQAGSKLASQLKQEGCEYVIALTHMRTPNDIKLAEGCPDIDLILGGHDHVYEVLEINNTYIVKSGTDFRQFSKISISFKGEAPKVEITEVPVTSSYPEDEVLKAKVEKYSAMIEGKMDEVLGKFSVPLEGRFSYIRRMECNLGNWVTDVLLAATGADLVVLNSGTFRSDQVHPAGDFTLRDLSTIIPMRDPLVVVEASGDVILQVLENSVSKYPSLEGRFPQVAGVSFAFDPSLPPGRRVAEHVVKVGDEYLQRAQRYRLAVKQYLRAGNDGYTMLPSCKLLVSEDMCPEIGMAIQNHFAAINVRTGKSKPSKHRQSLVTLSRRHSLVKMLDVNELDGPPPLRRASSAAVEPTPHSGHHRLTRRASLDDLEQQSCELAPKIENRIVVLDKPEKLQALLAERARWESDTVIREVEDENSP
Summary
Similarity
Belongs to the 5'-nucleotidase family.
Uniprot
H9JX74
A0A2W1BCW2
A0A3S2LQT4
A0A194QLC8
A0A2H1VC72
D6WST0
+ More
A0A1Y1LR40 A0A1Y1LR30 D2T1A9 A0A1B6DWA8 A0A067R593 A0A1Y1LSZ6 A0A2A4IY91 A0A1B6K7N1 A0A1B6CPL6 A0A0N0PB47 A0A1B6JJ67 A0A1B6LIG1 A0A1L8DXR8 A0A1L8DXW0 A0A182WIJ9 A0A182NHG7 U5EPQ2 A0A182JQV5 A0A1B0CJE1 B0X2W1 W5JTJ7 T1P8W7 A0A1I8N2J0 A0A1I8N2M0 A0A182R4R2 A0A0L0BPU7 A0A1Q3F4L9 A0A2M4BG11 A0A182FFY6 A0A1Q3F4I6 A0A2M4CZL5 A0A1Q3F6X0 A0A2M4BFQ8 A0A2M4BFT8 T1PK91 A0A1I8PAL9 Q5TX58 A0A1I8PAS0 A0A182XZJ2 A0A182H2E3 Q16MX9 Q16MX8 A0A2M4AMJ4 A0A2M4AM95 A0A1A9ZY03 J9JKF3 A0A2P8Z3L8 A0A2H8TKV1 A0A1B0C0P4 A0A2S2Q3W1 A0A1A9X3Q6 A0A2J7QM57 A0A1A9YI13 A0A1A9UKW9 A0A182PWM7 A0A1B0FAJ7 A0A2S2PGA5 A0A182X4U4 W8BA01 W8BF12 A0A1J1J4X5 A0A023F9K6 A0A0Q9XHK3 A0A0P4VX18 B4KN18 A0A0Q9X851 A0A0Q9X7Q3 B4K1T8 B4J7S3 A0A0A1XR45 B4QAL5 A0A0R3NPW4 B4GG78 B4HMS6 A0A0Q9W572 A0A0A1XLD1 A0A2M4BIX8 B4LKN0 A0A0Q9W5J9 A0A0M3QUH1 A0A0J9RAV3 A0A0R3NPL6 A0A1W4VPU7 A8DY95 A0A0Q9W524 A0A3B0JLL2 N6W6M4 A0A0R3NX19 A0A0J9TYF4 A0A3B0J019 A0A2J7QM06 A0A0J9R9T3 A1Z8A7 A0A1W4VQJ4
A0A1Y1LR40 A0A1Y1LR30 D2T1A9 A0A1B6DWA8 A0A067R593 A0A1Y1LSZ6 A0A2A4IY91 A0A1B6K7N1 A0A1B6CPL6 A0A0N0PB47 A0A1B6JJ67 A0A1B6LIG1 A0A1L8DXR8 A0A1L8DXW0 A0A182WIJ9 A0A182NHG7 U5EPQ2 A0A182JQV5 A0A1B0CJE1 B0X2W1 W5JTJ7 T1P8W7 A0A1I8N2J0 A0A1I8N2M0 A0A182R4R2 A0A0L0BPU7 A0A1Q3F4L9 A0A2M4BG11 A0A182FFY6 A0A1Q3F4I6 A0A2M4CZL5 A0A1Q3F6X0 A0A2M4BFQ8 A0A2M4BFT8 T1PK91 A0A1I8PAL9 Q5TX58 A0A1I8PAS0 A0A182XZJ2 A0A182H2E3 Q16MX9 Q16MX8 A0A2M4AMJ4 A0A2M4AM95 A0A1A9ZY03 J9JKF3 A0A2P8Z3L8 A0A2H8TKV1 A0A1B0C0P4 A0A2S2Q3W1 A0A1A9X3Q6 A0A2J7QM57 A0A1A9YI13 A0A1A9UKW9 A0A182PWM7 A0A1B0FAJ7 A0A2S2PGA5 A0A182X4U4 W8BA01 W8BF12 A0A1J1J4X5 A0A023F9K6 A0A0Q9XHK3 A0A0P4VX18 B4KN18 A0A0Q9X851 A0A0Q9X7Q3 B4K1T8 B4J7S3 A0A0A1XR45 B4QAL5 A0A0R3NPW4 B4GG78 B4HMS6 A0A0Q9W572 A0A0A1XLD1 A0A2M4BIX8 B4LKN0 A0A0Q9W5J9 A0A0M3QUH1 A0A0J9RAV3 A0A0R3NPL6 A0A1W4VPU7 A8DY95 A0A0Q9W524 A0A3B0JLL2 N6W6M4 A0A0R3NX19 A0A0J9TYF4 A0A3B0J019 A0A2J7QM06 A0A0J9R9T3 A1Z8A7 A0A1W4VQJ4
Pubmed
19121390
28756777
26354079
18362917
19820115
28004739
+ More
24845553 20920257 23761445 25315136 26108605 12364791 14747013 17210077 25244985 26483478 17510324 29403074 24495485 25474469 17994087 27129103 25830018 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
24845553 20920257 23761445 25315136 26108605 12364791 14747013 17210077 25244985 26483478 17510324 29403074 24495485 25474469 17994087 27129103 25830018 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01035854
BABH01035855
BABH01035856
KZ150152
PZC72818.1
RSAL01000405
+ More
RVE41893.1 KQ459053 KPJ04246.1 ODYU01001752 SOQ38386.1 KQ971352 EFA06359.1 GEZM01049101 JAV76103.1 GEZM01049102 JAV76102.1 FM986354 CAX20734.1 GEDC01007324 JAS29974.1 KK852686 KDR18455.1 GEZM01049103 JAV76101.1 NWSH01005099 PCG64436.1 GECU01000260 JAT07447.1 GEDC01021930 JAS15368.1 KQ461157 KPJ08087.1 GECU01008527 JAS99179.1 GEBQ01016593 JAT23384.1 GFDF01002841 JAV11243.1 GFDF01002840 JAV11244.1 GANO01004595 JAB55276.1 AJWK01014530 AJWK01014531 AJWK01014532 AJWK01014533 AJWK01014534 DS232301 EDS39435.1 ADMH02000483 ETN66280.1 KA645137 AFP59766.1 JRES01001567 KNC21993.1 GFDL01012572 JAV22473.1 GGFJ01002833 MBW51974.1 GFDL01012598 JAV22447.1 GGFL01006493 MBW70671.1 GFDL01011730 JAV23315.1 GGFJ01002744 MBW51885.1 GGFJ01002746 MBW51887.1 KA649114 AFP63743.1 AAAB01008807 EAL41709.2 JXUM01105151 JXUM01105152 KQ564943 KXJ71523.1 CH477846 EAT35695.1 EAT35696.1 GGFK01008695 MBW42016.1 GGFK01008578 MBW41899.1 ABLF02032200 PYGN01000212 PSN51090.1 GFXV01002477 MBW14282.1 JXJN01023713 JXJN01023714 GGMS01003204 MBY72407.1 NEVH01013220 PNF29638.1 CCAG010018439 GGMR01015852 MBY28471.1 GAMC01011063 JAB95492.1 GAMC01011061 GAMC01011059 GAMC01011052 GAMC01011046 JAB95503.1 CVRI01000067 CRL06860.1 GBBI01001109 JAC17603.1 CH933808 KRG04302.1 GDKW01001376 JAI55219.1 EDW08845.2 KRG04301.1 KRG04300.1 CH917851 EDV90856.1 CH916367 EDW01129.1 GBXI01000860 JAD13432.1 CM000362 EDX06520.1 CM000071 KRT03002.1 CH479183 EDW35498.1 CH480816 EDW47288.1 CH940648 KRF80218.1 GBXI01002944 JAD11348.1 GGFJ01003869 MBW53010.1 EDW61753.2 KRF80216.1 CP012524 ALC40619.1 CM002911 KMY92834.1 KRT03000.1 AE013599 ABV53755.1 AHN56087.1 KRF80217.1 OUUW01000001 SPP74136.1 ENO01863.2 KRT02998.1 KMY92835.1 SPP74134.1 PNF29639.1 KMY92833.1 AAF58767.1
RVE41893.1 KQ459053 KPJ04246.1 ODYU01001752 SOQ38386.1 KQ971352 EFA06359.1 GEZM01049101 JAV76103.1 GEZM01049102 JAV76102.1 FM986354 CAX20734.1 GEDC01007324 JAS29974.1 KK852686 KDR18455.1 GEZM01049103 JAV76101.1 NWSH01005099 PCG64436.1 GECU01000260 JAT07447.1 GEDC01021930 JAS15368.1 KQ461157 KPJ08087.1 GECU01008527 JAS99179.1 GEBQ01016593 JAT23384.1 GFDF01002841 JAV11243.1 GFDF01002840 JAV11244.1 GANO01004595 JAB55276.1 AJWK01014530 AJWK01014531 AJWK01014532 AJWK01014533 AJWK01014534 DS232301 EDS39435.1 ADMH02000483 ETN66280.1 KA645137 AFP59766.1 JRES01001567 KNC21993.1 GFDL01012572 JAV22473.1 GGFJ01002833 MBW51974.1 GFDL01012598 JAV22447.1 GGFL01006493 MBW70671.1 GFDL01011730 JAV23315.1 GGFJ01002744 MBW51885.1 GGFJ01002746 MBW51887.1 KA649114 AFP63743.1 AAAB01008807 EAL41709.2 JXUM01105151 JXUM01105152 KQ564943 KXJ71523.1 CH477846 EAT35695.1 EAT35696.1 GGFK01008695 MBW42016.1 GGFK01008578 MBW41899.1 ABLF02032200 PYGN01000212 PSN51090.1 GFXV01002477 MBW14282.1 JXJN01023713 JXJN01023714 GGMS01003204 MBY72407.1 NEVH01013220 PNF29638.1 CCAG010018439 GGMR01015852 MBY28471.1 GAMC01011063 JAB95492.1 GAMC01011061 GAMC01011059 GAMC01011052 GAMC01011046 JAB95503.1 CVRI01000067 CRL06860.1 GBBI01001109 JAC17603.1 CH933808 KRG04302.1 GDKW01001376 JAI55219.1 EDW08845.2 KRG04301.1 KRG04300.1 CH917851 EDV90856.1 CH916367 EDW01129.1 GBXI01000860 JAD13432.1 CM000362 EDX06520.1 CM000071 KRT03002.1 CH479183 EDW35498.1 CH480816 EDW47288.1 CH940648 KRF80218.1 GBXI01002944 JAD11348.1 GGFJ01003869 MBW53010.1 EDW61753.2 KRF80216.1 CP012524 ALC40619.1 CM002911 KMY92834.1 KRT03000.1 AE013599 ABV53755.1 AHN56087.1 KRF80217.1 OUUW01000001 SPP74136.1 ENO01863.2 KRT02998.1 KMY92835.1 SPP74134.1 PNF29639.1 KMY92833.1 AAF58767.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000007266
UP000027135
UP000218220
+ More
UP000053240 UP000075920 UP000075884 UP000075881 UP000092461 UP000002320 UP000000673 UP000095301 UP000075900 UP000037069 UP000069272 UP000095300 UP000007062 UP000076408 UP000069940 UP000249989 UP000008820 UP000092445 UP000007819 UP000245037 UP000092460 UP000091820 UP000235965 UP000092443 UP000078200 UP000075885 UP000092444 UP000076407 UP000183832 UP000009192 UP000001070 UP000000304 UP000001819 UP000008744 UP000001292 UP000008792 UP000092553 UP000192221 UP000000803 UP000268350
UP000053240 UP000075920 UP000075884 UP000075881 UP000092461 UP000002320 UP000000673 UP000095301 UP000075900 UP000037069 UP000069272 UP000095300 UP000007062 UP000076408 UP000069940 UP000249989 UP000008820 UP000092445 UP000007819 UP000245037 UP000092460 UP000091820 UP000235965 UP000092443 UP000078200 UP000075885 UP000092444 UP000076407 UP000183832 UP000009192 UP000001070 UP000000304 UP000001819 UP000008744 UP000001292 UP000008792 UP000092553 UP000192221 UP000000803 UP000268350
Pfam
Interpro
IPR008334
5'-Nucleotdase_C
+ More
IPR036907 5'-Nucleotdase_C_sf
IPR029052 Metallo-depent_PP-like
IPR004843 Calcineurin-like_PHP_ApaH
IPR041821 CG11883_N
IPR006179 5_nucleotidase/apyrase
IPR036291 NAD(P)-bd_dom_sf
IPR002347 SDR_fam
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR020472 G-protein_beta_WD-40_rep
IPR021772 WDR48/Bun107
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR019335 COG7
IPR001164 ArfGAP_dom
IPR022018 GIT1_C
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR038508 ArfGAP_dom_sf
IPR037278 ARFGAP/RecO
IPR002110 Ankyrin_rpt
IPR013724 GIT_SHD
IPR036397 RNaseH_sf
IPR002562 3'-5'_exonuclease_dom
IPR012337 RNaseH-like_sf
IPR036907 5'-Nucleotdase_C_sf
IPR029052 Metallo-depent_PP-like
IPR004843 Calcineurin-like_PHP_ApaH
IPR041821 CG11883_N
IPR006179 5_nucleotidase/apyrase
IPR036291 NAD(P)-bd_dom_sf
IPR002347 SDR_fam
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR020472 G-protein_beta_WD-40_rep
IPR021772 WDR48/Bun107
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR019335 COG7
IPR001164 ArfGAP_dom
IPR022018 GIT1_C
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR038508 ArfGAP_dom_sf
IPR037278 ARFGAP/RecO
IPR002110 Ankyrin_rpt
IPR013724 GIT_SHD
IPR036397 RNaseH_sf
IPR002562 3'-5'_exonuclease_dom
IPR012337 RNaseH-like_sf
SUPFAM
ProteinModelPortal
H9JX74
A0A2W1BCW2
A0A3S2LQT4
A0A194QLC8
A0A2H1VC72
D6WST0
+ More
A0A1Y1LR40 A0A1Y1LR30 D2T1A9 A0A1B6DWA8 A0A067R593 A0A1Y1LSZ6 A0A2A4IY91 A0A1B6K7N1 A0A1B6CPL6 A0A0N0PB47 A0A1B6JJ67 A0A1B6LIG1 A0A1L8DXR8 A0A1L8DXW0 A0A182WIJ9 A0A182NHG7 U5EPQ2 A0A182JQV5 A0A1B0CJE1 B0X2W1 W5JTJ7 T1P8W7 A0A1I8N2J0 A0A1I8N2M0 A0A182R4R2 A0A0L0BPU7 A0A1Q3F4L9 A0A2M4BG11 A0A182FFY6 A0A1Q3F4I6 A0A2M4CZL5 A0A1Q3F6X0 A0A2M4BFQ8 A0A2M4BFT8 T1PK91 A0A1I8PAL9 Q5TX58 A0A1I8PAS0 A0A182XZJ2 A0A182H2E3 Q16MX9 Q16MX8 A0A2M4AMJ4 A0A2M4AM95 A0A1A9ZY03 J9JKF3 A0A2P8Z3L8 A0A2H8TKV1 A0A1B0C0P4 A0A2S2Q3W1 A0A1A9X3Q6 A0A2J7QM57 A0A1A9YI13 A0A1A9UKW9 A0A182PWM7 A0A1B0FAJ7 A0A2S2PGA5 A0A182X4U4 W8BA01 W8BF12 A0A1J1J4X5 A0A023F9K6 A0A0Q9XHK3 A0A0P4VX18 B4KN18 A0A0Q9X851 A0A0Q9X7Q3 B4K1T8 B4J7S3 A0A0A1XR45 B4QAL5 A0A0R3NPW4 B4GG78 B4HMS6 A0A0Q9W572 A0A0A1XLD1 A0A2M4BIX8 B4LKN0 A0A0Q9W5J9 A0A0M3QUH1 A0A0J9RAV3 A0A0R3NPL6 A0A1W4VPU7 A8DY95 A0A0Q9W524 A0A3B0JLL2 N6W6M4 A0A0R3NX19 A0A0J9TYF4 A0A3B0J019 A0A2J7QM06 A0A0J9R9T3 A1Z8A7 A0A1W4VQJ4
A0A1Y1LR40 A0A1Y1LR30 D2T1A9 A0A1B6DWA8 A0A067R593 A0A1Y1LSZ6 A0A2A4IY91 A0A1B6K7N1 A0A1B6CPL6 A0A0N0PB47 A0A1B6JJ67 A0A1B6LIG1 A0A1L8DXR8 A0A1L8DXW0 A0A182WIJ9 A0A182NHG7 U5EPQ2 A0A182JQV5 A0A1B0CJE1 B0X2W1 W5JTJ7 T1P8W7 A0A1I8N2J0 A0A1I8N2M0 A0A182R4R2 A0A0L0BPU7 A0A1Q3F4L9 A0A2M4BG11 A0A182FFY6 A0A1Q3F4I6 A0A2M4CZL5 A0A1Q3F6X0 A0A2M4BFQ8 A0A2M4BFT8 T1PK91 A0A1I8PAL9 Q5TX58 A0A1I8PAS0 A0A182XZJ2 A0A182H2E3 Q16MX9 Q16MX8 A0A2M4AMJ4 A0A2M4AM95 A0A1A9ZY03 J9JKF3 A0A2P8Z3L8 A0A2H8TKV1 A0A1B0C0P4 A0A2S2Q3W1 A0A1A9X3Q6 A0A2J7QM57 A0A1A9YI13 A0A1A9UKW9 A0A182PWM7 A0A1B0FAJ7 A0A2S2PGA5 A0A182X4U4 W8BA01 W8BF12 A0A1J1J4X5 A0A023F9K6 A0A0Q9XHK3 A0A0P4VX18 B4KN18 A0A0Q9X851 A0A0Q9X7Q3 B4K1T8 B4J7S3 A0A0A1XR45 B4QAL5 A0A0R3NPW4 B4GG78 B4HMS6 A0A0Q9W572 A0A0A1XLD1 A0A2M4BIX8 B4LKN0 A0A0Q9W5J9 A0A0M3QUH1 A0A0J9RAV3 A0A0R3NPL6 A0A1W4VPU7 A8DY95 A0A0Q9W524 A0A3B0JLL2 N6W6M4 A0A0R3NX19 A0A0J9TYF4 A0A3B0J019 A0A2J7QM06 A0A0J9R9T3 A1Z8A7 A0A1W4VQJ4
PDB
2Z1A
E-value=9.68528e-37,
Score=387
Ontologies
PATHWAY
GO
PANTHER
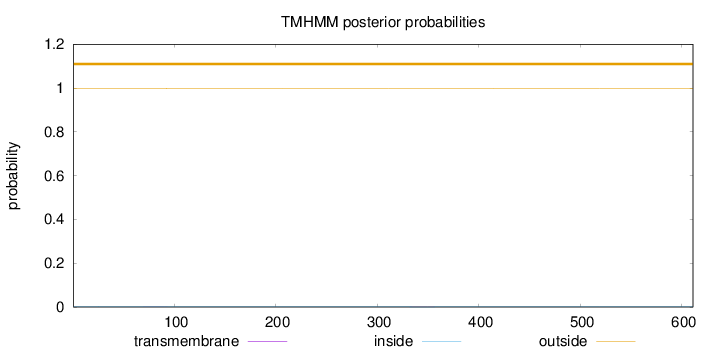
Topology
Length:
611
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02797
Exp number, first 60 AAs:
3e-05
Total prob of N-in:
0.00149
outside
1 - 611
Population Genetic Test Statistics
Pi
229.160907
Theta
156.065624
Tajima's D
1.927025
CLR
0.548031
CSRT
0.871506424678766
Interpretation
Uncertain
