Pre Gene Modal
BGIBMGA014140
Annotation
PREDICTED:_protein_LTV1_homolog_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.089
Sequence
CDS
ATGCCTAAAACAAAAAAGAAATTCATAGATAAAAAGAAAGCGGTGACGTTTAACCTAGTACACAGATCTCAAAGAGATCCTCTGGCCGCCGACGAAACAGCCCCGCAGCGAGTATTAGTGCCCGTAAACACAATCATCCCACCTACAAAGGATAAAGATACAGAATACACACCTGAACAACGCAAAGAGGAACAAAGGAAATACGGTATATATTACGACGATGACTACAATTATTTGCAGCATCTCAAAGACACAAAAGAGGTTACACTTGTGTTGCAGCCTAAAACTCCACATAAGAAGAAGCCAACGGAATCAACAAAAACTGATGAAATCATCGATGAAGATGAAGAGCCGTCCATTCCAGTCACAGAACGTCTGATGCTGCCCAGTTCGGTGTTCGCTTCTGAATTTGAAGAGGATGTTGGTTTACTCAATAAAGCTGCTCCACAGGGTTTACGCCTCGATCTGGATCCGGAAGTAGTGGCCGCTTTGGATGAAGATTTTGATTTTGATGATCCTGAAAATGAACTGGAAGACAACTTCATCGAATTGGCTATGGGAGACGATGATGAAGAAGCATATGATGAAGATGAAGGTTCATCGGGTGATGATAACTCTGAGAAAGCGTTCTCTTCAGACTTTGAATCAAATGATGAAAGCGATTCGCAGGATGAAGACAACCGCGGCAGGATGCCGACGTGGGCGCCGCCGGACAAGGACGACACCAAGTCGCGGTTCTCACAGTACTCGATGTCGTCCAGCATTATAAGGCGTAACCAGGGCTTGGAGCTACTGGACAACAGATTCGAAAAGATGTACGCCGAGTACGACGACACAGAGCTGGGTGCCCTCGACCTGGACGAGATCGAGGGTCACATGCCGGAGACGCACGCCATGCTGCTGCAGGCCGCCCAGGAGTTCGAGGCCTCGCAGAGGAAGTACAAGCTCGACAAGGAAACCGAAATGAGGAGGTTACAACGTCTTCAGGAAATCGAAGAAGAATCTGAAGAAGATGTCGTCACCGTGGAAGTTGAGCCCAAGGAGAGATGGGACTGCGAGACAATCCTCAGCACCTACTCCAACTTGTACAACCATCCGAAACTCATAGAGGAACCTAAGAAACCGAAGAGGATCGTGATAGATCCAAAAACTGGTATACCTAAGGATGTGCTCGGCACCGACGGTGGGAGGCTGACGGTGAAGGCGCTGGCCAGGTTCAACGCCATTAGCGAGGCCGCACCGCACGCCGCCGACGCCGCCAGCCGCGCCGACACGCTGCTGTCCACGCTCAGCGCGCTCTCCATAAGGCCCACAGACGAAAGTGTTGAAGAAAAGAGAGAACGAAAACGTTTATTGAAAGAATACAGACGAGAGAGGAGAATCGAGAAGAAAGCTAATAAAGAAGCTTTCAAGGAAGAGAAGAAACGGCAAGAGAAGATCATGATGAACAACAAGAATAATGTGCAAGGGAACAGAATATTGTAG
Protein
MPKTKKKFIDKKKAVTFNLVHRSQRDPLAADETAPQRVLVPVNTIIPPTKDKDTEYTPEQRKEEQRKYGIYYDDDYNYLQHLKDTKEVTLVLQPKTPHKKKPTESTKTDEIIDEDEEPSIPVTERLMLPSSVFASEFEEDVGLLNKAAPQGLRLDLDPEVVAALDEDFDFDDPENELEDNFIELAMGDDDEEAYDEDEGSSGDDNSEKAFSSDFESNDESDSQDEDNRGRMPTWAPPDKDDTKSRFSQYSMSSSIIRRNQGLELLDNRFEKMYAEYDDTELGALDLDEIEGHMPETHAMLLQAAQEFEASQRKYKLDKETEMRRLQRLQEIEEESEEDVVTVEVEPKERWDCETILSTYSNLYNHPKLIEEPKKPKRIVIDPKTGIPKDVLGTDGGRLTVKALARFNAISEAAPHAADAASRADTLLSTLSALSIRPTDESVEEKRERKRLLKEYRRERRIEKKANKEAFKEEKKRQEKIMMNNKNNVQGNRIL
Summary
Cofactor
FAD
Uniprot
H9JX75
A0A2W1BC16
A0A2A4IUR1
A0A194QLB8
A0A0N0PB50
A0A1E1WGT3
+ More
A0A212F4H7 A0A212ESW0 A0A2Z5U870 A0A0L7RE50 A0A0J7NML6 A0A026WQA4 D6WQU3 A0A336K3U7 A0A1Y1L875 E9IUT4 A0A158NZ60 A0A154PCV9 E2A666 A0A195AZ41 A0A336LKM3 A0A195F631 F4X357 A0A182YA34 A0A1S4H4D6 Q7PPS6 A0A182SDE6 B0XJL7 A0A182JRK4 A0A182X1U9 A0A182UY65 A0A182LQ23 A0A182TPU4 A0A182HFR3 A0A182M3R0 K7IUB3 A0A182VTB9 A0A232FER1 A0A023EVX4 A0A023ET53 A0A1Q3F9N9 U5EY17 A0A182P8H2 Q17MQ1 A0A182RLP6 A0A182MYD7 A0A182ITA9 A0A182R1E6 A0A226F1P4 A0A2P8Z569 C1BUM5 A0A195CVZ8
A0A212F4H7 A0A212ESW0 A0A2Z5U870 A0A0L7RE50 A0A0J7NML6 A0A026WQA4 D6WQU3 A0A336K3U7 A0A1Y1L875 E9IUT4 A0A158NZ60 A0A154PCV9 E2A666 A0A195AZ41 A0A336LKM3 A0A195F631 F4X357 A0A182YA34 A0A1S4H4D6 Q7PPS6 A0A182SDE6 B0XJL7 A0A182JRK4 A0A182X1U9 A0A182UY65 A0A182LQ23 A0A182TPU4 A0A182HFR3 A0A182M3R0 K7IUB3 A0A182VTB9 A0A232FER1 A0A023EVX4 A0A023ET53 A0A1Q3F9N9 U5EY17 A0A182P8H2 Q17MQ1 A0A182RLP6 A0A182MYD7 A0A182ITA9 A0A182R1E6 A0A226F1P4 A0A2P8Z569 C1BUM5 A0A195CVZ8
Pubmed
EMBL
BABH01035849
BABH01035850
BABH01035851
KZ150152
PZC72829.1
NWSH01007296
+ More
PCG63014.1 KQ459053 KPJ04236.1 KQ461157 KPJ08097.1 GDQN01004983 JAT86071.1 AGBW02010359 OWR48641.1 AGBW02012726 OWR44534.1 AP017505 BBB06810.1 KQ414613 KOC69001.1 LBMM01003266 KMQ93730.1 KK107151 QOIP01000003 EZA57294.1 RLU24326.1 KQ971354 EFA07437.1 UFQS01000039 UFQT01000039 SSW98125.1 SSX18511.1 GEZM01062351 JAV69794.1 GL766050 EFZ15661.1 ADTU01003909 KQ434864 KZC09098.1 GL437108 EFN71125.1 KQ976701 KYM77307.1 SSW98124.1 SSX18510.1 KQ981768 KYN35928.1 GL888608 EGI59157.1 AAAB01008933 EAA09915.4 DS233552 EDS30515.1 APCN01005567 AXCM01002293 AAZX01010653 NNAY01000357 OXU28929.1 GAPW01001134 JAC12464.1 GAPW01001135 JAC12463.1 GFDL01010827 JAV24218.1 GANO01000609 JAB59262.1 CH477204 EAT47971.1 AXCN02001048 LNIX01000001 OXA63705.1 PYGN01000190 PSN51643.1 BT078304 ACO12728.1 KQ977220 KYN04858.1
PCG63014.1 KQ459053 KPJ04236.1 KQ461157 KPJ08097.1 GDQN01004983 JAT86071.1 AGBW02010359 OWR48641.1 AGBW02012726 OWR44534.1 AP017505 BBB06810.1 KQ414613 KOC69001.1 LBMM01003266 KMQ93730.1 KK107151 QOIP01000003 EZA57294.1 RLU24326.1 KQ971354 EFA07437.1 UFQS01000039 UFQT01000039 SSW98125.1 SSX18511.1 GEZM01062351 JAV69794.1 GL766050 EFZ15661.1 ADTU01003909 KQ434864 KZC09098.1 GL437108 EFN71125.1 KQ976701 KYM77307.1 SSW98124.1 SSX18510.1 KQ981768 KYN35928.1 GL888608 EGI59157.1 AAAB01008933 EAA09915.4 DS233552 EDS30515.1 APCN01005567 AXCM01002293 AAZX01010653 NNAY01000357 OXU28929.1 GAPW01001134 JAC12464.1 GAPW01001135 JAC12463.1 GFDL01010827 JAV24218.1 GANO01000609 JAB59262.1 CH477204 EAT47971.1 AXCN02001048 LNIX01000001 OXA63705.1 PYGN01000190 PSN51643.1 BT078304 ACO12728.1 KQ977220 KYN04858.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000053825
+ More
UP000036403 UP000053097 UP000279307 UP000007266 UP000005205 UP000076502 UP000000311 UP000078540 UP000078541 UP000007755 UP000076408 UP000007062 UP000075901 UP000002320 UP000075881 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000075883 UP000002358 UP000075920 UP000215335 UP000075885 UP000008820 UP000075900 UP000075884 UP000075880 UP000075886 UP000198287 UP000245037 UP000078542
UP000036403 UP000053097 UP000279307 UP000007266 UP000005205 UP000076502 UP000000311 UP000078540 UP000078541 UP000007755 UP000076408 UP000007062 UP000075901 UP000002320 UP000075881 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000075883 UP000002358 UP000075920 UP000215335 UP000075885 UP000008820 UP000075900 UP000075884 UP000075880 UP000075886 UP000198287 UP000245037 UP000078542
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JX75
A0A2W1BC16
A0A2A4IUR1
A0A194QLB8
A0A0N0PB50
A0A1E1WGT3
+ More
A0A212F4H7 A0A212ESW0 A0A2Z5U870 A0A0L7RE50 A0A0J7NML6 A0A026WQA4 D6WQU3 A0A336K3U7 A0A1Y1L875 E9IUT4 A0A158NZ60 A0A154PCV9 E2A666 A0A195AZ41 A0A336LKM3 A0A195F631 F4X357 A0A182YA34 A0A1S4H4D6 Q7PPS6 A0A182SDE6 B0XJL7 A0A182JRK4 A0A182X1U9 A0A182UY65 A0A182LQ23 A0A182TPU4 A0A182HFR3 A0A182M3R0 K7IUB3 A0A182VTB9 A0A232FER1 A0A023EVX4 A0A023ET53 A0A1Q3F9N9 U5EY17 A0A182P8H2 Q17MQ1 A0A182RLP6 A0A182MYD7 A0A182ITA9 A0A182R1E6 A0A226F1P4 A0A2P8Z569 C1BUM5 A0A195CVZ8
A0A212F4H7 A0A212ESW0 A0A2Z5U870 A0A0L7RE50 A0A0J7NML6 A0A026WQA4 D6WQU3 A0A336K3U7 A0A1Y1L875 E9IUT4 A0A158NZ60 A0A154PCV9 E2A666 A0A195AZ41 A0A336LKM3 A0A195F631 F4X357 A0A182YA34 A0A1S4H4D6 Q7PPS6 A0A182SDE6 B0XJL7 A0A182JRK4 A0A182X1U9 A0A182UY65 A0A182LQ23 A0A182TPU4 A0A182HFR3 A0A182M3R0 K7IUB3 A0A182VTB9 A0A232FER1 A0A023EVX4 A0A023ET53 A0A1Q3F9N9 U5EY17 A0A182P8H2 Q17MQ1 A0A182RLP6 A0A182MYD7 A0A182ITA9 A0A182R1E6 A0A226F1P4 A0A2P8Z569 C1BUM5 A0A195CVZ8
PDB
6G53
E-value=4.55192e-28,
Score=311
Ontologies
PANTHER
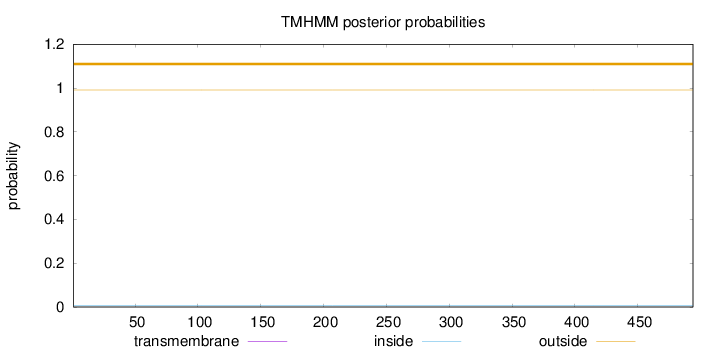
Topology
Length:
494
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00851
outside
1 - 494
Population Genetic Test Statistics
Pi
289.286519
Theta
180.671672
Tajima's D
2.19762
CLR
0.412396
CSRT
0.909054547272636
Interpretation
Uncertain
