Gene
KWMTBOMO12241 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014194
Annotation
PREDICTED:_vacuolar_protein_sorting-associated_protein_35_isoform_X1_[Bombyx_mori]
Full name
Vacuolar protein sorting-associated protein 35
Alternative Name
Maternal-embryonic 3
Vesicle protein sorting 35
Vesicle protein sorting 35
Location in the cell
Cytoplasmic Reliability : 1.607 Nuclear Reliability : 1.021
Sequence
CDS
ATGAAGATCATCAAGCTGCCCTGCGACCACTACAAGAACATACTGGTGCTGATTAAGTTGTCCAATTACGCGCCGCTGATACGACATCTCAGCTACCAGGGTCGGGTGATGATAGCCGTCCACCTCATCAACGACGTGCTGGAGACCAACACCGCAATATCCACTCCGCAAGATGTCGAAGCTGTCCTCGGTCTCTTGGATGTGCTGGTGAAGGATCAGCCGGATCAGCCGCCTTCTGAAAGCGACGTTGAAGACTTCGTGGAAGAACAGGGCTTGTTGGCCAGACTTATTCATCATTTCAAATCGGACTCCGCCGACCAGCAGTATCTGATAACCAGCACAGCGAGGAAGATCCTTCAGGGCGGCGGAGCGAGGAGGATCCAGCACACGTTCCCGCCGTTGGTGTTCCACGCGTACCGTCTGGCGTTCGTTTATAAGGACCTCAAAGACCAGGACGAGATGTGGGAGAAGAAATGCCAGAAGATATTCCAGTTCTGTCACCAGACCATCACCATGCTCGTGAAGGCGGAACTGGCTGAACTCCCTCTAAGGTTGTACCTCCAGGGAGCTTTGGCGATAAGCGAAATAGGATTTGCCAATCATGAAACGATAGCTTACGAGTTCCTGTCGCAAGCTTTCTCGCTGTATGAAGACGAGATATCCGACAGCAAAGCGCAACTGGCCGCAATAACATTGATAATAGCCACCTTCGAGCAGATCAATTGTTTTGGCGCTGAGAACGCGGAGCCGATGCGGACCCAGTGCGCGCTGGCGGCCAGCAAGCTGCTGAAGAAGCCGGACCAGAGCAGGGCCGTGGCGCTCTGCTCGCATCTCTTCTGGAAGGGGACCAGGGACGGCCGCCCGGTGTGGGCGCTGAACGACGCGTCCCGCGCGCTGGACTGCCTGAAGAAGGCGGCCCGCGTGGCCCAGCAGTGCATGGACGGCGGCGTGCAGGCGCAGCTGCTGGCCGAGCTGCTGGGCCGATACGCGCTGCTGCGGGAGCGCGGCAACCAGAGTCTCACGCCCAGCCTCATCGACGCGATAATACAGAAAATACGCGAGGAGCTGGCCAACTTAGATCAGTCCGAAGAAGTGGAGCAGATCACGAAACACTTCCACAACACTTTGCAGCATCTGAAGAACAGGATCGAGTCTCCGGACCCCGAGAGTCTGGGCTACGAGGGTCTCACGCTGTCCTAA
Protein
MKIIKLPCDHYKNILVLIKLSNYAPLIRHLSYQGRVMIAVHLINDVLETNTAISTPQDVEAVLGLLDVLVKDQPDQPPSESDVEDFVEEQGLLARLIHHFKSDSADQQYLITSTARKILQGGGARRIQHTFPPLVFHAYRLAFVYKDLKDQDEMWEKKCQKIFQFCHQTITMLVKAELAELPLRLYLQGALAISEIGFANHETIAYEFLSQAFSLYEDEISDSKAQLAAITLIIATFEQINCFGAENAEPMRTQCALAASKLLKKPDQSRAVALCSHLFWKGTRDGRPVWALNDASRALDCLKKAARVAQQCMDGGVQAQLLAELLGRYALLRERGNQSLTPSLIDAIIQKIREELANLDQSEEVEQITKHFHNTLQHLKNRIESPDPESLGYEGLTLS
Summary
Description
Plays a role in vesicular protein sorting.
Acts as component of the retromer cargo-selective complex (CSC). The CSC is believed to be the core functional component of retromer or respective retromer complex variants acting to prevent missorting of selected transmembrane cargo proteins into the lysosomal degradation pathway. The recruitment of the CSC to the endosomal membrane involves RAB7A and SNX3. The CSC seems to associate with the cytoplasmic domain of cargo proteins predominantly via VPS35; however, these interactions seem to be of low affinity and retromer SNX proteins may also contribute to cargo selectivity thus questioning the classical function of the CSC. The SNX-BAR retromer mediates retrograde transport of cargo proteins from endosomes to the trans-Golgi network (TGN) and is involved in endosome-to-plasma membrane transport for cargo protein recycling. The SNX3-retromer mediates the retrograde transport of WLS distinct from the SNX-BAR retromer pathway. The SNX27-retromer is believed to be involved in endosome-to-plasma membrane trafficking and recycling of a broad spectrum of cargo proteins. The CSC seems to act as recruitment hub for other proteins, such as the WASH complex and TBC1D5 (Probable). Required for retrograde transport of lysosomal enzyme receptor IGF2R and SLC11A2. Required to regulate transcytosis of the polymeric immunoglobulin receptor (pIgR-pIgA). Required for endosomal localization of WASHC2 and mediates the association of the CSC with the WASH complex (By similarity).
Acts as component of the retromer cargo-selective complex (CSC). The CSC is believed to be the core functional component of retromer or respective retromer complex variants acting to prevent missorting of selected transmembrane cargo proteins into the lysosomal degradation pathway. The recruitment of the CSC to the endosomal membrane involves RAB7A and SNX3. The CSC seems to associate with the cytoplasmic domain of cargo proteins predominantly via VPS35; however, these interactions seem to be of low affinity and retromer SNX proteins may also contribute to cargo selectivity thus questioning the classical function of the CSC. The SNX-BAR retromer mediates retrograde transport of cargo proteins from endosomes to the trans-Golgi network (TGN) and is involved in endosome-to-plasma membrane transport for cargo protein recycling. The SNX3-retromer mediates the retrograde transport of WLS distinct from the SNX-BAR retromer pathway. The SNX27-retromer is believed to be involved in endosome-to-plasma membrane trafficking and recycling of a broad spectrum of cargo proteins. The CSC seems to act as recruitment hub for other proteins, such as the WASH complex and TBC1D5 (Probable). Required for retrograde transport of lysosomal enzyme receptor IGF2R and SLC11A2. Required to regulate transcytosis of the polymeric immunoglobulin receptor (pIgR-pIgA). Required for endosomal localization of WASHC2 and mediates the association of the CSC with the WASH complex (By similarity).
Subunit
Component of the heterotrimeric retromer cargo-selective complex (CSC), also described as vacuolar protein sorting subcomplex (VPS) formed by VPS26 (VPS26A or VPS26B), VPS29 and VPS35. The CSC has a highly elongated structure with VPS26 and VPS29 binding independently at opposite distal ends of VPS35 as central platform (PubMed:21040701, PubMed:20875039, PubMed:21920005). The CSC is believed to associate with variable sorting nexins to form functionally distinct retromer complex variants. The originally described retromer complex (also called SNX-BAR retromer) is a pentamer containing the CSC and a heterodimeric membrane-deforming subcomplex formed between SNX1 or SNX2 and SNX5 or SNX6 (also called SNX-BAR subcomplex); the affinity between the respective CSC and SNX-BAR subcomplexes is low. The CSC associates with SNX3 to form a SNX3-retromer complex. The CSC associates with SNX27, the WASH complex and the SNX-BAR subcomplex to form the SNX27-retromer complex (Probable). Interacts with VPS26A, VPS29, VPS26B and LRRK2 (PubMed:16190980, PubMed:21040701, PubMed:20875039, PubMed:21920005, PubMed:23395371). Interacts with SNX1, SNX2, IGF2R, SNX3, GOLPH3, SLC11A2, WASHC2, FKBP15, WASHC1, EHD1. Interacts with MAGEL2; leading to recruitment of the TRIM27:MAGEL2 E3 ubiquitin ligase complex retromer-containing endosomes (By similarity).
Similarity
Belongs to the VPS35 family.
Keywords
Complete proteome
Cytoplasm
Direct protein sequencing
Endosome
Membrane
Phosphoprotein
Protein transport
Reference proteome
Transport
Feature
chain Vacuolar protein sorting-associated protein 35
Uniprot
A0A194QFH4
A0A1E1WPM0
A0A2A4J3P7
H9JXC9
A0A0N0PAV2
A0A2Z5U7Y2
+ More
A0A2W1BHY5 A0A1B6BWC4 A0A1Y1L5K3 A0A1Y1L104 A0A154PEM6 A0A026X171 A0A3L8DR35 A0A1B6JB37 A0A0L7RH01 A0A0J7KZE6 A0A0N0BFA1 A0A1B6LFS4 F4WW33 A0A310SSW8 A0A195B1Z9 A0A158NLJ4 A0A195CRL7 A0A195E754 A0A2J7RBJ3 A0A195EYX3 A0A151XKC0 A0A0C9R1A1 A0A0C9RRG2 A0A0C9Q717 E2BGH6 A0A067REE6 A0A0T6B773 A0A087ZYP1 E2B0Q3 A0A2A3EEH7 K7ITP4 A0A093ESR9 C0PU95 A0A091L8E6 A0A2K6D3M2 A0A2K5WX53 A0A2I3T974 A0A1W4WZP8 I7GPC4 A0A2U3Z435 B5DFC1 K9KEJ7 V9K8I6 A4QP42 Q1ED30 A0A3Q0DXW8 A0A3P9AEM0 A0A2I0M2I5 A0A3L7HZ98 Q6ZM34 A0A061IC10 A0A3P8YVD1 A0A3B4G4Z7 A0A3P8PEY6 Q3TKU6 A0A3B3B7F9 A0A3S2N731 W5UH12 A0A2D0Q7L1 A0A091J4C6 A0A091FL01 Q3TRJ1 Q9EQH3 A0A1U7S401 A0A2I3MLH5 A0A093HKA9 A0A218UWD5 A0A1V4KPU8 R0JL21 A0A3L8S0K6 A0A093PHN3 A0A091UDZ5 A0A091JUI9 A0A099ZZN4 A0A093NWN7 A0A091RD27 A0A087QX31 H0ZF25 A0A060VUG2 W5LGY1 A0A099YWH3 A0A091NKN6 A0A091F1P6 A0A093CT46 A0A3B4G2N2 U3JBP3 A0A093QUD4 A0A1U7R5B8 A0A091TA01 A0A091V0H4 A0A2Y9M0G7 A0A341BYZ5 A0A2U4B467 A0A3Q3C0N9
A0A2W1BHY5 A0A1B6BWC4 A0A1Y1L5K3 A0A1Y1L104 A0A154PEM6 A0A026X171 A0A3L8DR35 A0A1B6JB37 A0A0L7RH01 A0A0J7KZE6 A0A0N0BFA1 A0A1B6LFS4 F4WW33 A0A310SSW8 A0A195B1Z9 A0A158NLJ4 A0A195CRL7 A0A195E754 A0A2J7RBJ3 A0A195EYX3 A0A151XKC0 A0A0C9R1A1 A0A0C9RRG2 A0A0C9Q717 E2BGH6 A0A067REE6 A0A0T6B773 A0A087ZYP1 E2B0Q3 A0A2A3EEH7 K7ITP4 A0A093ESR9 C0PU95 A0A091L8E6 A0A2K6D3M2 A0A2K5WX53 A0A2I3T974 A0A1W4WZP8 I7GPC4 A0A2U3Z435 B5DFC1 K9KEJ7 V9K8I6 A4QP42 Q1ED30 A0A3Q0DXW8 A0A3P9AEM0 A0A2I0M2I5 A0A3L7HZ98 Q6ZM34 A0A061IC10 A0A3P8YVD1 A0A3B4G4Z7 A0A3P8PEY6 Q3TKU6 A0A3B3B7F9 A0A3S2N731 W5UH12 A0A2D0Q7L1 A0A091J4C6 A0A091FL01 Q3TRJ1 Q9EQH3 A0A1U7S401 A0A2I3MLH5 A0A093HKA9 A0A218UWD5 A0A1V4KPU8 R0JL21 A0A3L8S0K6 A0A093PHN3 A0A091UDZ5 A0A091JUI9 A0A099ZZN4 A0A093NWN7 A0A091RD27 A0A087QX31 H0ZF25 A0A060VUG2 W5LGY1 A0A099YWH3 A0A091NKN6 A0A091F1P6 A0A093CT46 A0A3B4G2N2 U3JBP3 A0A093QUD4 A0A1U7R5B8 A0A091TA01 A0A091V0H4 A0A2Y9M0G7 A0A341BYZ5 A0A2U4B467 A0A3Q3C0N9
Pubmed
26354079
19121390
28756777
28004739
24508170
30249741
+ More
21719571 21347285 20798317 24845553 20075255 16136131 17194215 15489334 24402279 25069045 23371554 29704459 23594743 23929341 10349636 11042159 11076861 11217851 12466851 16141073 29451363 23127152 12040188 17947660 19131326 21183079 8678978 11112353 16190980 21040701 20875039 21920005 23395371 30282656 20360741 24755649 25329095
21719571 21347285 20798317 24845553 20075255 16136131 17194215 15489334 24402279 25069045 23371554 29704459 23594743 23929341 10349636 11042159 11076861 11217851 12466851 16141073 29451363 23127152 12040188 17947660 19131326 21183079 8678978 11112353 16190980 21040701 20875039 21920005 23395371 30282656 20360741 24755649 25329095
EMBL
KQ459053
KPJ04237.1
GDQN01002197
JAT88857.1
NWSH01003436
PCG66308.1
+ More
BABH01035847 BABH01035848 BABH01035849 KQ461157 KPJ08096.1 AP017506 BBB06811.1 KZ150152 PZC72827.1 GEDC01031715 JAS05583.1 GEZM01067994 JAV67275.1 GEZM01067995 JAV67274.1 KQ434878 KZC09884.1 KK107063 EZA61104.1 QOIP01000005 RLU22742.1 GECU01011358 GECU01008054 JAS96348.1 JAS99652.1 KQ414596 KOC70113.1 LBMM01001796 KMQ95736.1 KQ435804 KOX73031.1 GEBQ01017448 GEBQ01009775 JAT22529.1 JAT30202.1 GL888404 EGI61622.1 KQ759820 OAD62720.1 KQ976662 KYM78315.1 ADTU01019551 KQ977349 KYN03351.1 KQ979579 KYN20669.1 NEVH01005896 PNF38206.1 KQ981914 KYN33107.1 KQ982038 KYQ60787.1 GBYB01009924 JAG79691.1 GBYB01009923 GBYB01009926 JAG79690.1 JAG79693.1 GBYB01009928 JAG79695.1 GL448189 EFN85169.1 KK852686 KDR18449.1 LJIG01009391 KRT83174.1 GL444666 EFN60725.1 KZ288269 PBC29874.1 KK615337 KFV48357.1 BT072172 ACN58635.1 KL302788 KFP51548.1 AQIA01042781 AQIA01042782 AACZ04030068 AB174124 BAE91186.1 BC169004 AAI69004.1 JL641145 AEQ00924.1 JW861487 AFO94004.1 BC139644 AAI39645.1 BC117573 AAI17574.1 AKCR02000046 PKK23890.1 RAZU01000127 RLQ71266.1 AL928906 KE672984 ERE78825.1 AK166821 BAE39047.1 CM012439 RVE74984.1 JT410194 AHH38962.1 KL218569 KFP06661.1 KL447078 KFO69631.1 AK150682 AK162726 AK167421 AK168880 AK170473 CH466525 BAE29762.1 BAE37038.1 BAE39509.1 BAE40698.1 BAE41818.1 EDL11004.1 U47024 AF226323 BC005469 BC006637 AHZZ02018658 AHZZ02018659 AHZZ02018660 AHZZ02018661 AHZZ02018662 KL206526 KFV83103.1 MUZQ01000107 OWK58124.1 LSYS01002285 OPJ86464.1 KB743571 EOA97990.1 QUSF01000110 RLV91534.1 KL669378 KFW75941.1 KK430718 KFQ88944.1 KK501485 KFP15261.1 KL870252 KGL87266.1 KL224832 KFW64537.1 KK813735 KFQ37386.1 KL225962 KFM05785.1 ABQF01034926 FR904258 CDQ55990.1 KL885851 KGL73293.1 KL384598 KFP89464.1 KK719362 KFO63161.1 KL465352 KFV16064.1 AGTO01010909 KL433035 KFW92131.1 KK442340 KFQ70600.1 KL410289 KFQ95770.1
BABH01035847 BABH01035848 BABH01035849 KQ461157 KPJ08096.1 AP017506 BBB06811.1 KZ150152 PZC72827.1 GEDC01031715 JAS05583.1 GEZM01067994 JAV67275.1 GEZM01067995 JAV67274.1 KQ434878 KZC09884.1 KK107063 EZA61104.1 QOIP01000005 RLU22742.1 GECU01011358 GECU01008054 JAS96348.1 JAS99652.1 KQ414596 KOC70113.1 LBMM01001796 KMQ95736.1 KQ435804 KOX73031.1 GEBQ01017448 GEBQ01009775 JAT22529.1 JAT30202.1 GL888404 EGI61622.1 KQ759820 OAD62720.1 KQ976662 KYM78315.1 ADTU01019551 KQ977349 KYN03351.1 KQ979579 KYN20669.1 NEVH01005896 PNF38206.1 KQ981914 KYN33107.1 KQ982038 KYQ60787.1 GBYB01009924 JAG79691.1 GBYB01009923 GBYB01009926 JAG79690.1 JAG79693.1 GBYB01009928 JAG79695.1 GL448189 EFN85169.1 KK852686 KDR18449.1 LJIG01009391 KRT83174.1 GL444666 EFN60725.1 KZ288269 PBC29874.1 KK615337 KFV48357.1 BT072172 ACN58635.1 KL302788 KFP51548.1 AQIA01042781 AQIA01042782 AACZ04030068 AB174124 BAE91186.1 BC169004 AAI69004.1 JL641145 AEQ00924.1 JW861487 AFO94004.1 BC139644 AAI39645.1 BC117573 AAI17574.1 AKCR02000046 PKK23890.1 RAZU01000127 RLQ71266.1 AL928906 KE672984 ERE78825.1 AK166821 BAE39047.1 CM012439 RVE74984.1 JT410194 AHH38962.1 KL218569 KFP06661.1 KL447078 KFO69631.1 AK150682 AK162726 AK167421 AK168880 AK170473 CH466525 BAE29762.1 BAE37038.1 BAE39509.1 BAE40698.1 BAE41818.1 EDL11004.1 U47024 AF226323 BC005469 BC006637 AHZZ02018658 AHZZ02018659 AHZZ02018660 AHZZ02018661 AHZZ02018662 KL206526 KFV83103.1 MUZQ01000107 OWK58124.1 LSYS01002285 OPJ86464.1 KB743571 EOA97990.1 QUSF01000110 RLV91534.1 KL669378 KFW75941.1 KK430718 KFQ88944.1 KK501485 KFP15261.1 KL870252 KGL87266.1 KL224832 KFW64537.1 KK813735 KFQ37386.1 KL225962 KFM05785.1 ABQF01034926 FR904258 CDQ55990.1 KL885851 KGL73293.1 KL384598 KFP89464.1 KK719362 KFO63161.1 KL465352 KFV16064.1 AGTO01010909 KL433035 KFW92131.1 KK442340 KFQ70600.1 KL410289 KFQ95770.1
Proteomes
UP000053268
UP000218220
UP000005204
UP000053240
UP000076502
UP000053097
+ More
UP000279307 UP000053825 UP000036403 UP000053105 UP000007755 UP000078540 UP000005205 UP000078542 UP000078492 UP000235965 UP000078541 UP000075809 UP000008237 UP000027135 UP000005203 UP000000311 UP000242457 UP000002358 UP000233120 UP000233100 UP000002277 UP000192223 UP000245341 UP000189704 UP000265140 UP000053872 UP000273346 UP000000437 UP000030759 UP000261460 UP000265100 UP000261560 UP000221080 UP000054308 UP000053760 UP000000589 UP000189705 UP000028761 UP000053584 UP000197619 UP000190648 UP000276834 UP000053258 UP000053119 UP000053858 UP000054081 UP000053286 UP000007754 UP000193380 UP000018467 UP000053641 UP000052976 UP000016665 UP000189706 UP000053283 UP000248483 UP000252040 UP000245320 UP000264840
UP000279307 UP000053825 UP000036403 UP000053105 UP000007755 UP000078540 UP000005205 UP000078542 UP000078492 UP000235965 UP000078541 UP000075809 UP000008237 UP000027135 UP000005203 UP000000311 UP000242457 UP000002358 UP000233120 UP000233100 UP000002277 UP000192223 UP000245341 UP000189704 UP000265140 UP000053872 UP000273346 UP000000437 UP000030759 UP000261460 UP000265100 UP000261560 UP000221080 UP000054308 UP000053760 UP000000589 UP000189705 UP000028761 UP000053584 UP000197619 UP000190648 UP000276834 UP000053258 UP000053119 UP000053858 UP000054081 UP000053286 UP000007754 UP000193380 UP000018467 UP000053641 UP000052976 UP000016665 UP000189706 UP000053283 UP000248483 UP000252040 UP000245320 UP000264840
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
A0A194QFH4
A0A1E1WPM0
A0A2A4J3P7
H9JXC9
A0A0N0PAV2
A0A2Z5U7Y2
+ More
A0A2W1BHY5 A0A1B6BWC4 A0A1Y1L5K3 A0A1Y1L104 A0A154PEM6 A0A026X171 A0A3L8DR35 A0A1B6JB37 A0A0L7RH01 A0A0J7KZE6 A0A0N0BFA1 A0A1B6LFS4 F4WW33 A0A310SSW8 A0A195B1Z9 A0A158NLJ4 A0A195CRL7 A0A195E754 A0A2J7RBJ3 A0A195EYX3 A0A151XKC0 A0A0C9R1A1 A0A0C9RRG2 A0A0C9Q717 E2BGH6 A0A067REE6 A0A0T6B773 A0A087ZYP1 E2B0Q3 A0A2A3EEH7 K7ITP4 A0A093ESR9 C0PU95 A0A091L8E6 A0A2K6D3M2 A0A2K5WX53 A0A2I3T974 A0A1W4WZP8 I7GPC4 A0A2U3Z435 B5DFC1 K9KEJ7 V9K8I6 A4QP42 Q1ED30 A0A3Q0DXW8 A0A3P9AEM0 A0A2I0M2I5 A0A3L7HZ98 Q6ZM34 A0A061IC10 A0A3P8YVD1 A0A3B4G4Z7 A0A3P8PEY6 Q3TKU6 A0A3B3B7F9 A0A3S2N731 W5UH12 A0A2D0Q7L1 A0A091J4C6 A0A091FL01 Q3TRJ1 Q9EQH3 A0A1U7S401 A0A2I3MLH5 A0A093HKA9 A0A218UWD5 A0A1V4KPU8 R0JL21 A0A3L8S0K6 A0A093PHN3 A0A091UDZ5 A0A091JUI9 A0A099ZZN4 A0A093NWN7 A0A091RD27 A0A087QX31 H0ZF25 A0A060VUG2 W5LGY1 A0A099YWH3 A0A091NKN6 A0A091F1P6 A0A093CT46 A0A3B4G2N2 U3JBP3 A0A093QUD4 A0A1U7R5B8 A0A091TA01 A0A091V0H4 A0A2Y9M0G7 A0A341BYZ5 A0A2U4B467 A0A3Q3C0N9
A0A2W1BHY5 A0A1B6BWC4 A0A1Y1L5K3 A0A1Y1L104 A0A154PEM6 A0A026X171 A0A3L8DR35 A0A1B6JB37 A0A0L7RH01 A0A0J7KZE6 A0A0N0BFA1 A0A1B6LFS4 F4WW33 A0A310SSW8 A0A195B1Z9 A0A158NLJ4 A0A195CRL7 A0A195E754 A0A2J7RBJ3 A0A195EYX3 A0A151XKC0 A0A0C9R1A1 A0A0C9RRG2 A0A0C9Q717 E2BGH6 A0A067REE6 A0A0T6B773 A0A087ZYP1 E2B0Q3 A0A2A3EEH7 K7ITP4 A0A093ESR9 C0PU95 A0A091L8E6 A0A2K6D3M2 A0A2K5WX53 A0A2I3T974 A0A1W4WZP8 I7GPC4 A0A2U3Z435 B5DFC1 K9KEJ7 V9K8I6 A4QP42 Q1ED30 A0A3Q0DXW8 A0A3P9AEM0 A0A2I0M2I5 A0A3L7HZ98 Q6ZM34 A0A061IC10 A0A3P8YVD1 A0A3B4G4Z7 A0A3P8PEY6 Q3TKU6 A0A3B3B7F9 A0A3S2N731 W5UH12 A0A2D0Q7L1 A0A091J4C6 A0A091FL01 Q3TRJ1 Q9EQH3 A0A1U7S401 A0A2I3MLH5 A0A093HKA9 A0A218UWD5 A0A1V4KPU8 R0JL21 A0A3L8S0K6 A0A093PHN3 A0A091UDZ5 A0A091JUI9 A0A099ZZN4 A0A093NWN7 A0A091RD27 A0A087QX31 H0ZF25 A0A060VUG2 W5LGY1 A0A099YWH3 A0A091NKN6 A0A091F1P6 A0A093CT46 A0A3B4G2N2 U3JBP3 A0A093QUD4 A0A1U7R5B8 A0A091TA01 A0A091V0H4 A0A2Y9M0G7 A0A341BYZ5 A0A2U4B467 A0A3Q3C0N9
PDB
5OSI
E-value=2.9695e-99,
Score=924
Ontologies
GO
GO:0005829
GO:0042147
GO:0030906
GO:0015031
GO:0016021
GO:0006886
GO:0005770
GO:0030904
GO:0010628
GO:0005769
GO:0097422
GO:0010008
GO:0005764
GO:2000331
GO:0031748
GO:1901215
GO:0099639
GO:0050728
GO:1902823
GO:0005739
GO:0099074
GO:0045056
GO:1903364
GO:0036010
GO:0043653
GO:0031647
GO:0090141
GO:0005886
GO:0060161
GO:1902950
GO:0099073
GO:0010629
GO:1990126
GO:0098978
GO:1903828
GO:0006624
GO:0032463
GO:0007040
GO:0098793
GO:0099003
GO:0048471
GO:1905166
GO:0014069
GO:1903181
GO:0043005
GO:0090263
GO:0098887
GO:0007416
GO:0061357
GO:0005768
GO:0050882
GO:0010821
GO:0033365
GO:0098794
GO:0043025
GO:0060548
GO:0005737
GO:0090326
GO:0031648
GO:0032268
GO:0008565
GO:0006428
GO:0004812
GO:0000049
GO:0005975
GO:0004553
PANTHER
Topology
Subcellular location
Cytoplasm
Membrane
Endosome
Early endosome
Late endosome
Membrane
Endosome
Early endosome
Late endosome
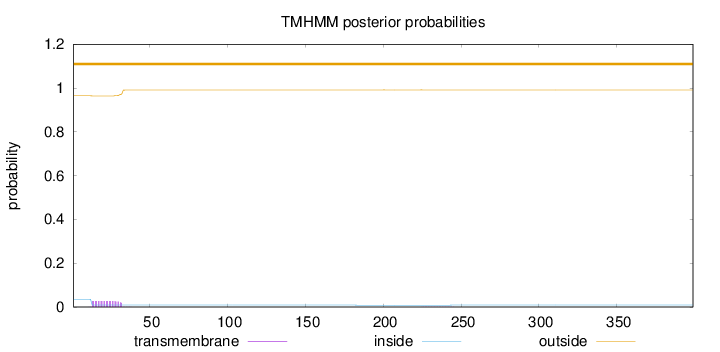
Length:
399
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.536520000000001
Exp number, first 60 AAs:
0.52703
Total prob of N-in:
0.03572
outside
1 - 399
Population Genetic Test Statistics
Pi
188.535129
Theta
139.584759
Tajima's D
1.481
CLR
0.136577
CSRT
0.783560821958902
Interpretation
Uncertain
