Gene
KWMTBOMO12240 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014194
Annotation
PREDICTED:_vacuolar_protein_sorting-associated_protein_35_isoform_X1_[Bombyx_mori]
Full name
Vacuolar protein sorting-associated protein 35
Location in the cell
Cytoplasmic Reliability : 1.802 Nuclear Reliability : 2.099
Sequence
CDS
ATGACAAACCAGGTGTCTCCAGTCGAAGAGCAGGAGAAACTCCTGGAGGAGGCCTTGAACATTGTTAAAGTTCAGGCATTTCAAATGAAAAGATGTCTCGACAAATCTAAATTAATGGACGCCCTGAAACATGCATCAACAATGCTGGGTGAATTGAGAACTTCACTGTTATCACCAAAGAGCTATTATGAGCTTTATATGGCAATAACAGATGAACTGCGTCACCTGGAGCTGTATCTTCTGGAGGAGTTCCAAAAGGGAAGGAAGGTTGCAGATCTTTATGAACTGGTCCAATATGCTGGGAACATTGTACCTAGATTGTACCTGTTAATCACAGTAGGACTGGTCTACATCAAAACAAACACGAATCTGCGAAGAGATCTATTGAAGGATCTGGTTGAGATGTGTCGCGGAGTTCAGCATCCTTTGAGAGGACTTTTCCTTAGGAATTACCTATTGCAGTGCACAAGGAACGTTTTACCGGACACACACGAAGCCCCAAATGAAAACGAGGGAACTGTACGAGATGCTGTTGATTTTGTCCTGATGAATTTCGCCGAAATGAACAAGCTGTGGGTCAGGATGCAGCACCAGGGACACTCAAGAGATAAAGAGCGCCGCGAACGAGAACGCTCAGAGCTTCGTATATTAGTTGGTACGAATTTGGTTCGACTGTCGCAACTGGAGTCCGTCACGGTGGAGGACTACCGCAGGATGGTGCTGCCCGGCATCTTGGAGCAGGTCGTGAGCTGCAGGGACGCCATAGCGCAGGAGTACCTCATGGAGTGTATCATACAGGTGTTCCCGGACGAGTTCCACCTGGCCAACTTACAGCCGTTCCTCAAGTCGTGTGCTGAACTCCAACCCGGAGTCAACATCAAGAACATCATCATAGCACTGATCGAGAGGCTCGCTTCGTATAGCCAGAGGAATGAAGGCAATATGAATCTAAGCGTGGTCCTGGAAGATGGACAAGAACAAGAAGTCCAACTGTTCGAAGTTTTCTCTGATCAAGTTGCTGCGATAACCCAGGTTAGCCAATCTTTGAAATACCCAGATATCATTTTAAAACAATGCAATGGCCGGCACAGATGTTACACAATCAATTGA
Protein
MTNQVSPVEEQEKLLEEALNIVKVQAFQMKRCLDKSKLMDALKHASTMLGELRTSLLSPKSYYELYMAITDELRHLELYLLEEFQKGRKVADLYELVQYAGNIVPRLYLLITVGLVYIKTNTNLRRDLLKDLVEMCRGVQHPLRGLFLRNYLLQCTRNVLPDTHEAPNENEGTVRDAVDFVLMNFAEMNKLWVRMQHQGHSRDKERRERERSELRILVGTNLVRLSQLESVTVEDYRRMVLPGILEQVVSCRDAIAQEYLMECIIQVFPDEFHLANLQPFLKSCAELQPGVNIKNIIIALIERLASYSQRNEGNMNLSVVLEDGQEQEVQLFEVFSDQVAAITQVSQSLKYPDIILKQCNGRHRCYTIN
Summary
Description
Plays a role in vesicular protein sorting.
Similarity
Belongs to the VPS35 family.
Uniprot
A0A1E1WPM0
A0A3S2L9C1
A0A2A4J3P7
A0A0N1INF0
A0A194QFH4
A0A2W1BHY5
+ More
H9JXC9 A0A1B6DHE0 A0A1B6DJ82 A0A2J7RBJ3 A0A067REE6 A0A1W4WZP8 A0A1Y1L5K3 A0A026X171 A0A154PEM6 A0A3L8DR35 A0A1Y1L104 A0A195CRL7 A0A0J7KZE6 K7ITP4 A0A195E754 A0A158NLJ4 A0A087ZYP1 A0A195B1Z9 A0A0C9R1A1 A0A0C9RRG2 A0A0C9Q717 A0A1B6G8I6 A0A1B6LFS4 A0A310SSW8 D6WTN7 A0A151XKC0 E2BGH6 E2B0Q3 A0A2A3EEH7 A0A195EYX3 E0W0D0 E9J671 A0A2H1WYA3 A0A2L2Y3Z6 A0A0N0BFA1 A0A1S3CUV1 A0A1B0C9T1 Q6DJN1 A0A1L8DVU9 F6STR3 A0A182WIW9 I7GI50 A0A084VF17 A0A182NHE2 A0A182TEI6 A0A2M4BE42 Q7PT27 W5JTI6 A0A182LWU4 A0A1S4FYT1 A0A3P4PKD1 Q16JB7 A0A182P9Z0 A0A182QYG6 A0A182FFX6 A0A182I3U4 A0A182X6J5 A0A182RB34 A0A2M4AIZ2 A0A0S7GMM4 A0A182J117 A0A182XZE0 A0A182SH88 A0A2K6KZ95 A0A2K6FCI8 T1ISA6 A0A1C7CZ67 A0A1D5Q1P9 A0A182JPG0 A0A182H4A9 A0A1Z5L3E2 A0A0S7GIP4 A0A2K5M5L6 A0A2K6D3R0 A0A2K6QZC5 A0A2I3LUV9 A0A2I3S6J6 A0A091SE65 A0A2I2YQ49 A0A1Q3F334 A0A1Q3F467 A0A090XCR5 A0A182VMW6 B3DL72 A0A2I3HK91 A0A1L8GLA7 A0A293LTE6 A0A146XV91 A0A093CT46 W5ME32 U4UI68 A0A218UWD5 A0A1V4KPU8 A0A093QUD4
H9JXC9 A0A1B6DHE0 A0A1B6DJ82 A0A2J7RBJ3 A0A067REE6 A0A1W4WZP8 A0A1Y1L5K3 A0A026X171 A0A154PEM6 A0A3L8DR35 A0A1Y1L104 A0A195CRL7 A0A0J7KZE6 K7ITP4 A0A195E754 A0A158NLJ4 A0A087ZYP1 A0A195B1Z9 A0A0C9R1A1 A0A0C9RRG2 A0A0C9Q717 A0A1B6G8I6 A0A1B6LFS4 A0A310SSW8 D6WTN7 A0A151XKC0 E2BGH6 E2B0Q3 A0A2A3EEH7 A0A195EYX3 E0W0D0 E9J671 A0A2H1WYA3 A0A2L2Y3Z6 A0A0N0BFA1 A0A1S3CUV1 A0A1B0C9T1 Q6DJN1 A0A1L8DVU9 F6STR3 A0A182WIW9 I7GI50 A0A084VF17 A0A182NHE2 A0A182TEI6 A0A2M4BE42 Q7PT27 W5JTI6 A0A182LWU4 A0A1S4FYT1 A0A3P4PKD1 Q16JB7 A0A182P9Z0 A0A182QYG6 A0A182FFX6 A0A182I3U4 A0A182X6J5 A0A182RB34 A0A2M4AIZ2 A0A0S7GMM4 A0A182J117 A0A182XZE0 A0A182SH88 A0A2K6KZ95 A0A2K6FCI8 T1ISA6 A0A1C7CZ67 A0A1D5Q1P9 A0A182JPG0 A0A182H4A9 A0A1Z5L3E2 A0A0S7GIP4 A0A2K5M5L6 A0A2K6D3R0 A0A2K6QZC5 A0A2I3LUV9 A0A2I3S6J6 A0A091SE65 A0A2I2YQ49 A0A1Q3F334 A0A1Q3F467 A0A090XCR5 A0A182VMW6 B3DL72 A0A2I3HK91 A0A1L8GLA7 A0A293LTE6 A0A146XV91 A0A093CT46 W5ME32 U4UI68 A0A218UWD5 A0A1V4KPU8 A0A093QUD4
Pubmed
26354079
28756777
19121390
24845553
28004739
24508170
+ More
30249741 20075255 21347285 18362917 19820115 20798317 20566863 21282665 26561354 18464734 17194215 24438588 12364791 14747013 17210077 20920257 23761445 17510324 25244985 20966253 17431167 26483478 28528879 25362486 16136131 22398555 25970599 27762356 23537049
30249741 20075255 21347285 18362917 19820115 20798317 20566863 21282665 26561354 18464734 17194215 24438588 12364791 14747013 17210077 20920257 23761445 17510324 25244985 20966253 17431167 26483478 28528879 25362486 16136131 22398555 25970599 27762356 23537049
EMBL
GDQN01002197
JAT88857.1
RSAL01000405
RVE41887.1
NWSH01003436
PCG66308.1
+ More
KQ461157 KPJ08095.1 KQ459053 KPJ04237.1 KZ150152 PZC72827.1 BABH01035847 BABH01035848 BABH01035849 GEDC01012177 JAS25121.1 GEDC01011537 JAS25761.1 NEVH01005896 PNF38206.1 KK852686 KDR18449.1 GEZM01067994 JAV67275.1 KK107063 EZA61104.1 KQ434878 KZC09884.1 QOIP01000005 RLU22742.1 GEZM01067995 JAV67274.1 KQ977349 KYN03351.1 LBMM01001796 KMQ95736.1 KQ979579 KYN20669.1 ADTU01019551 KQ976662 KYM78315.1 GBYB01009924 JAG79691.1 GBYB01009923 GBYB01009926 JAG79690.1 JAG79693.1 GBYB01009928 JAG79695.1 GECZ01011058 JAS58711.1 GEBQ01017448 GEBQ01009775 JAT22529.1 JAT30202.1 KQ759820 OAD62720.1 KQ971355 EFA05820.1 KQ982038 KYQ60787.1 GL448189 EFN85169.1 GL444666 EFN60725.1 KZ288269 PBC29874.1 KQ981914 KYN33107.1 DS235858 EEB19086.1 GL768221 EFZ11749.1 ODYU01011993 SOQ58055.1 IAAA01004349 IAAA01004350 IAAA01004351 IAAA01004352 IAAA01004353 LAA02902.1 KQ435804 KOX73031.1 AJWK01002738 BC075144 AAH75144.1 GFDF01003526 JAV10558.1 AB172209 BAE89271.1 ATLV01012360 KE524785 KFB36561.1 GGFJ01002174 MBW51315.1 AAAB01008807 EAA04647.5 ADMH02000483 ETN66270.1 AXCM01001132 CYRY02033022 VCX10288.1 CH478018 EAT34367.1 AXCN02000591 APCN01000222 GGFK01007429 MBW40750.1 GBYX01452493 JAO29022.1 JH431429 JSUE03026135 JXUM01109152 JXUM01109153 JXUM01109154 JXUM01109155 KQ565294 KXJ71121.1 GFJQ02005058 JAW01912.1 GBYX01452490 JAO29025.1 AHZZ02018658 AHZZ02018659 AHZZ02018660 AHZZ02018661 AHZZ02018662 AACZ04030068 KK948439 KFQ56315.1 CABD030100887 CABD030100888 GFDL01013137 JAV21908.1 GFDL01012683 JAV22362.1 GBIH01001580 JAC93130.1 BC167336 AAI67336.1 ADFV01124297 CM004472 OCT84624.1 GFWV01012018 GFWV01013508 MAA36747.1 GCES01040400 JAR45923.1 KL465352 KFV16064.1 AHAT01034963 KB632173 ERL89580.1 MUZQ01000107 OWK58124.1 LSYS01002285 OPJ86464.1 KL433035 KFW92131.1
KQ461157 KPJ08095.1 KQ459053 KPJ04237.1 KZ150152 PZC72827.1 BABH01035847 BABH01035848 BABH01035849 GEDC01012177 JAS25121.1 GEDC01011537 JAS25761.1 NEVH01005896 PNF38206.1 KK852686 KDR18449.1 GEZM01067994 JAV67275.1 KK107063 EZA61104.1 KQ434878 KZC09884.1 QOIP01000005 RLU22742.1 GEZM01067995 JAV67274.1 KQ977349 KYN03351.1 LBMM01001796 KMQ95736.1 KQ979579 KYN20669.1 ADTU01019551 KQ976662 KYM78315.1 GBYB01009924 JAG79691.1 GBYB01009923 GBYB01009926 JAG79690.1 JAG79693.1 GBYB01009928 JAG79695.1 GECZ01011058 JAS58711.1 GEBQ01017448 GEBQ01009775 JAT22529.1 JAT30202.1 KQ759820 OAD62720.1 KQ971355 EFA05820.1 KQ982038 KYQ60787.1 GL448189 EFN85169.1 GL444666 EFN60725.1 KZ288269 PBC29874.1 KQ981914 KYN33107.1 DS235858 EEB19086.1 GL768221 EFZ11749.1 ODYU01011993 SOQ58055.1 IAAA01004349 IAAA01004350 IAAA01004351 IAAA01004352 IAAA01004353 LAA02902.1 KQ435804 KOX73031.1 AJWK01002738 BC075144 AAH75144.1 GFDF01003526 JAV10558.1 AB172209 BAE89271.1 ATLV01012360 KE524785 KFB36561.1 GGFJ01002174 MBW51315.1 AAAB01008807 EAA04647.5 ADMH02000483 ETN66270.1 AXCM01001132 CYRY02033022 VCX10288.1 CH478018 EAT34367.1 AXCN02000591 APCN01000222 GGFK01007429 MBW40750.1 GBYX01452493 JAO29022.1 JH431429 JSUE03026135 JXUM01109152 JXUM01109153 JXUM01109154 JXUM01109155 KQ565294 KXJ71121.1 GFJQ02005058 JAW01912.1 GBYX01452490 JAO29025.1 AHZZ02018658 AHZZ02018659 AHZZ02018660 AHZZ02018661 AHZZ02018662 AACZ04030068 KK948439 KFQ56315.1 CABD030100887 CABD030100888 GFDL01013137 JAV21908.1 GFDL01012683 JAV22362.1 GBIH01001580 JAC93130.1 BC167336 AAI67336.1 ADFV01124297 CM004472 OCT84624.1 GFWV01012018 GFWV01013508 MAA36747.1 GCES01040400 JAR45923.1 KL465352 KFV16064.1 AHAT01034963 KB632173 ERL89580.1 MUZQ01000107 OWK58124.1 LSYS01002285 OPJ86464.1 KL433035 KFW92131.1
Proteomes
UP000283053
UP000218220
UP000053240
UP000053268
UP000005204
UP000235965
+ More
UP000027135 UP000192223 UP000053097 UP000076502 UP000279307 UP000078542 UP000036403 UP000002358 UP000078492 UP000005205 UP000005203 UP000078540 UP000007266 UP000075809 UP000008237 UP000000311 UP000242457 UP000078541 UP000009046 UP000053105 UP000079169 UP000092461 UP000002279 UP000075920 UP000030765 UP000075884 UP000075902 UP000007062 UP000000673 UP000075883 UP000008820 UP000075885 UP000075886 UP000069272 UP000075840 UP000076407 UP000075900 UP000075880 UP000076408 UP000075901 UP000233180 UP000233160 UP000075882 UP000006718 UP000075881 UP000069940 UP000249989 UP000233060 UP000233120 UP000233200 UP000028761 UP000002277 UP000001519 UP000075903 UP000001073 UP000186698 UP000018468 UP000030742 UP000197619 UP000190648
UP000027135 UP000192223 UP000053097 UP000076502 UP000279307 UP000078542 UP000036403 UP000002358 UP000078492 UP000005205 UP000005203 UP000078540 UP000007266 UP000075809 UP000008237 UP000000311 UP000242457 UP000078541 UP000009046 UP000053105 UP000079169 UP000092461 UP000002279 UP000075920 UP000030765 UP000075884 UP000075902 UP000007062 UP000000673 UP000075883 UP000008820 UP000075885 UP000075886 UP000069272 UP000075840 UP000076407 UP000075900 UP000075880 UP000076408 UP000075901 UP000233180 UP000233160 UP000075882 UP000006718 UP000075881 UP000069940 UP000249989 UP000233060 UP000233120 UP000233200 UP000028761 UP000002277 UP000001519 UP000075903 UP000001073 UP000186698 UP000018468 UP000030742 UP000197619 UP000190648
PRIDE
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
A0A1E1WPM0
A0A3S2L9C1
A0A2A4J3P7
A0A0N1INF0
A0A194QFH4
A0A2W1BHY5
+ More
H9JXC9 A0A1B6DHE0 A0A1B6DJ82 A0A2J7RBJ3 A0A067REE6 A0A1W4WZP8 A0A1Y1L5K3 A0A026X171 A0A154PEM6 A0A3L8DR35 A0A1Y1L104 A0A195CRL7 A0A0J7KZE6 K7ITP4 A0A195E754 A0A158NLJ4 A0A087ZYP1 A0A195B1Z9 A0A0C9R1A1 A0A0C9RRG2 A0A0C9Q717 A0A1B6G8I6 A0A1B6LFS4 A0A310SSW8 D6WTN7 A0A151XKC0 E2BGH6 E2B0Q3 A0A2A3EEH7 A0A195EYX3 E0W0D0 E9J671 A0A2H1WYA3 A0A2L2Y3Z6 A0A0N0BFA1 A0A1S3CUV1 A0A1B0C9T1 Q6DJN1 A0A1L8DVU9 F6STR3 A0A182WIW9 I7GI50 A0A084VF17 A0A182NHE2 A0A182TEI6 A0A2M4BE42 Q7PT27 W5JTI6 A0A182LWU4 A0A1S4FYT1 A0A3P4PKD1 Q16JB7 A0A182P9Z0 A0A182QYG6 A0A182FFX6 A0A182I3U4 A0A182X6J5 A0A182RB34 A0A2M4AIZ2 A0A0S7GMM4 A0A182J117 A0A182XZE0 A0A182SH88 A0A2K6KZ95 A0A2K6FCI8 T1ISA6 A0A1C7CZ67 A0A1D5Q1P9 A0A182JPG0 A0A182H4A9 A0A1Z5L3E2 A0A0S7GIP4 A0A2K5M5L6 A0A2K6D3R0 A0A2K6QZC5 A0A2I3LUV9 A0A2I3S6J6 A0A091SE65 A0A2I2YQ49 A0A1Q3F334 A0A1Q3F467 A0A090XCR5 A0A182VMW6 B3DL72 A0A2I3HK91 A0A1L8GLA7 A0A293LTE6 A0A146XV91 A0A093CT46 W5ME32 U4UI68 A0A218UWD5 A0A1V4KPU8 A0A093QUD4
H9JXC9 A0A1B6DHE0 A0A1B6DJ82 A0A2J7RBJ3 A0A067REE6 A0A1W4WZP8 A0A1Y1L5K3 A0A026X171 A0A154PEM6 A0A3L8DR35 A0A1Y1L104 A0A195CRL7 A0A0J7KZE6 K7ITP4 A0A195E754 A0A158NLJ4 A0A087ZYP1 A0A195B1Z9 A0A0C9R1A1 A0A0C9RRG2 A0A0C9Q717 A0A1B6G8I6 A0A1B6LFS4 A0A310SSW8 D6WTN7 A0A151XKC0 E2BGH6 E2B0Q3 A0A2A3EEH7 A0A195EYX3 E0W0D0 E9J671 A0A2H1WYA3 A0A2L2Y3Z6 A0A0N0BFA1 A0A1S3CUV1 A0A1B0C9T1 Q6DJN1 A0A1L8DVU9 F6STR3 A0A182WIW9 I7GI50 A0A084VF17 A0A182NHE2 A0A182TEI6 A0A2M4BE42 Q7PT27 W5JTI6 A0A182LWU4 A0A1S4FYT1 A0A3P4PKD1 Q16JB7 A0A182P9Z0 A0A182QYG6 A0A182FFX6 A0A182I3U4 A0A182X6J5 A0A182RB34 A0A2M4AIZ2 A0A0S7GMM4 A0A182J117 A0A182XZE0 A0A182SH88 A0A2K6KZ95 A0A2K6FCI8 T1ISA6 A0A1C7CZ67 A0A1D5Q1P9 A0A182JPG0 A0A182H4A9 A0A1Z5L3E2 A0A0S7GIP4 A0A2K5M5L6 A0A2K6D3R0 A0A2K6QZC5 A0A2I3LUV9 A0A2I3S6J6 A0A091SE65 A0A2I2YQ49 A0A1Q3F334 A0A1Q3F467 A0A090XCR5 A0A182VMW6 B3DL72 A0A2I3HK91 A0A1L8GLA7 A0A293LTE6 A0A146XV91 A0A093CT46 W5ME32 U4UI68 A0A218UWD5 A0A1V4KPU8 A0A093QUD4
PDB
5F0P
E-value=2.75458e-124,
Score=1139
Ontologies
GO
PANTHER
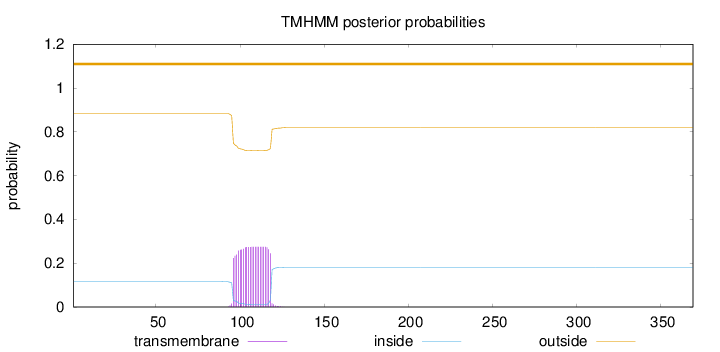
Topology
Length:
369
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.17081999999999
Exp number, first 60 AAs:
0.00141
Total prob of N-in:
0.11622
outside
1 - 369
Population Genetic Test Statistics
Pi
257.12203
Theta
180.672821
Tajima's D
1.258244
CLR
0
CSRT
0.729413529323534
Interpretation
Uncertain
