Gene
KWMTBOMO12239
Pre Gene Modal
BGIBMGA014142
Annotation
PREDICTED:_trafficking_protein_particle_complex_subunit_12_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.979
Sequence
CDS
ATGGATAATAAACCGTCATTGAGTCAATATTTCGGAGGATCAGAGATACCACCAGCTTCTCAGTTCTTCGACGAAATCGGAACATCGCCATCAGATATGATACAAAGCGTTTATTTAGGTGAAAGCGAAGGGTCTCGAACAACAGCACCCAGTGTTTTCACGTCAAGTACGTCAGCAACATTTTCTCAACCGTCCACTCTTGATGAAGTCACTTTTTCAAATGTTGTACCCACTGTGAGTGGGACCACATCTCTACCTGCCACCAGCTTGCCTGATCCATCAACGTTTTTCGATACAATCGGCCCTGAACCTACTATAGGGCTGACAAAACCTGTAACAACTACAGCACTGACTGATGCTATGGACGGATTGAGTATTAAGAATCAACCTGTAGACAAAGATGCAGAAAGAAGACGGGACGCTTGGATACCCAATGAAGAAGCCAAGAAGACATTGATGAAGGCACAATCGTCACCTAAAGGTTCATTTTTCCCTGAGAGAGAAGTGTTGACCATGCCAGGATTGGTGCTTGAAGAAGAACTGGCGGACGCGTTACAAGAGGTGGCGGTGAAGTACCTGGGCGTGAGCTGCGCGGGCAGCCGCGGGGTGGTGCGCGCCGACCACGTGACCCGGGACGAGCCCGGCCTCCGCGAGCTCATCCGCACCGGGTACTTCAGGGCGGCCGTCAACCTCACCGCGGTGCTCATCAGCGCCGCCGGACAGGGCGTGGGACGGATGCACAGACCCACGAAGCACACGCCGCGCTCCCTCCAGCTCTGGTTGACGAGGTTCTCCCTGATGCTGAGGATCAAGCTACATGAACCCCTGATCAGGGAGGCGGAGCAGTTCGGCGACTGCACCAAACCCGACATGTTTTACCAGTTCTACCCGGAGTCTTACGAGAACCGTCCGGGATCCCTCGTTCCTTTCTCGCTGAGGCTACTCCTGGCCGAGATACCGGGGCACGTGGGCAAGCCGGACGAGTCCATGGACAGACTATACGCGATGCTCGATATTATACAGCAGATGCTGTCGAATCTCCGCGACGGAAAGACCGAGGACGGCTCTGGTCTCCTCTCGTCGGAGCAGGATAAGGCGGAGTCGATACGTTTGTGGATGGGGAGGGAGACGAGGGTGTATCATTCGATCGTCAACTGCGCTATAGCCCTGAAGGATTACAGGCACGCTTCCAACGTGCTGTCCTCGTTGGAACAGGCGTGCAGCACGGCCGGTCAGAAGCGAGCCGTGTGCAGCGCGCTATGCCGCCTCGCGCTGCTGGCCGGGCACGTGACGGCCGCACGTGCGCACTGCCGCCGCGCCACCGAGAACCGGACGCACGTATGTCCGACGCCCGACGTGCGCGAGTACGTGGACCTCGGGCTGGTGGCGATGGCTCAGGGGAAGTACGAAGACGCCTACAACGACTTCGTGAGGGCGGCCGACCAGGAACCCACGAACATTATGGTCGCGAACAACATCTCCGTGTGCCTGTTGTACATGGGTCGCCTGAAGGAGGCCATATCGGTGCTGCAGAAGGCGATAAAGAGTGACCCGGAACGCGGCCTCAACGAGAGCCTCCTCGTCAACCTGTGCACGCTTTACGAACTGGAGTCGTCCAAGACGAACGAGAAAAAGCTCAATCTGTTGCGGATGCTGTGCAAGTACAAGAGCGACACGATACCGAACGTTCTCGAATGTCTGAAGCTCGCGTAG
Protein
MDNKPSLSQYFGGSEIPPASQFFDEIGTSPSDMIQSVYLGESEGSRTTAPSVFTSSTSATFSQPSTLDEVTFSNVVPTVSGTTSLPATSLPDPSTFFDTIGPEPTIGLTKPVTTTALTDAMDGLSIKNQPVDKDAERRRDAWIPNEEAKKTLMKAQSSPKGSFFPEREVLTMPGLVLEEELADALQEVAVKYLGVSCAGSRGVVRADHVTRDEPGLRELIRTGYFRAAVNLTAVLISAAGQGVGRMHRPTKHTPRSLQLWLTRFSLMLRIKLHEPLIREAEQFGDCTKPDMFYQFYPESYENRPGSLVPFSLRLLLAEIPGHVGKPDESMDRLYAMLDIIQQMLSNLRDGKTEDGSGLLSSEQDKAESIRLWMGRETRVYHSIVNCAIALKDYRHASNVLSSLEQACSTAGQKRAVCSALCRLALLAGHVTAARAHCRRATENRTHVCPTPDVREYVDLGLVAMAQGKYEDAYNDFVRAADQEPTNIMVANNISVCLLYMGRLKEAISVLQKAIKSDPERGLNESLLVNLCTLYELESSKTNEKKLNLLRMLCKYKSDTIPNVLECLKLA
Summary
Uniprot
A0A2H1V0P3
A0A2W1BJ11
A0A2A4J360
A0A194QFB7
A0A212FIJ7
A0A0N0PCP1
+ More
V5GCW3 D6WPG0 U4UQG0 N6TWI2 A0A1Y1KKS6 A0A1W4XHY4 A0A1B6BYH9 A0A2J7PS11 A0A2J7PS04 A0A2J7PS14 A0A2R7WXT7 T1HMT7 A0A1B6KVC4 A0A067R6H4 A0A0V0G9Y9 A0A069DUF9 A0A224XBV9 A0A0K8S371 A0A0A9XAH5 A0A0P4VNY7 A0A026VX52 A0A146KLC0 A0A146LLA5 A0A3L8DGQ1 A0A1B6I8Y1 E2ABT3 A0A154PEY2 A0A0N7ZC98 A0A0J7NGY9 A0A0P4W5J3 A0A0P4WC21 E9IED2 A0A195BZH9 A0A158NP13 E2BS49 A0A195DX02 A0A310S5J5 A0A195EZZ2 F4W8X7 A0A0C9QWN8 A0A232FJ39 A0A151ILD2 A0A2A3E2T3 A0A151X8F6 K7J170 A0A088ADV5 E0VWB0 T1J077 A0A1B0DCJ2 A0A2R5L4M2 A0A182IW73 A0A1B6M3G4 A0A1I8N0H4 A0A293M422 K1PZM5 A0A2M4AN58 A0A1Z5KVB0 A0A2M4AN87 A0A1I8N0J4 T1PJB0 A0A1E1XBL9 A0A0L0CJW7 A0A1E1XL09 A0A131XXQ9 A0A1I8PVJ0 A0A1I8PVM2 W5L350 A0A226F2K3 V5GZ85 A0A1A9WDI6 A0A182H370 A0A1S3IL77 A0A3B4DRX5 A0A1D2NIX9 A0A210QAJ3 A0A224YKU9 A0A131YCY1 Q16HT7 A0A1S4G0D1 L7M0X7 A0A131YW63 W5JU93 A0A182W2G0 U4UHS3 A0A1B0AD56 A0A182K617 N6TYS3
V5GCW3 D6WPG0 U4UQG0 N6TWI2 A0A1Y1KKS6 A0A1W4XHY4 A0A1B6BYH9 A0A2J7PS11 A0A2J7PS04 A0A2J7PS14 A0A2R7WXT7 T1HMT7 A0A1B6KVC4 A0A067R6H4 A0A0V0G9Y9 A0A069DUF9 A0A224XBV9 A0A0K8S371 A0A0A9XAH5 A0A0P4VNY7 A0A026VX52 A0A146KLC0 A0A146LLA5 A0A3L8DGQ1 A0A1B6I8Y1 E2ABT3 A0A154PEY2 A0A0N7ZC98 A0A0J7NGY9 A0A0P4W5J3 A0A0P4WC21 E9IED2 A0A195BZH9 A0A158NP13 E2BS49 A0A195DX02 A0A310S5J5 A0A195EZZ2 F4W8X7 A0A0C9QWN8 A0A232FJ39 A0A151ILD2 A0A2A3E2T3 A0A151X8F6 K7J170 A0A088ADV5 E0VWB0 T1J077 A0A1B0DCJ2 A0A2R5L4M2 A0A182IW73 A0A1B6M3G4 A0A1I8N0H4 A0A293M422 K1PZM5 A0A2M4AN58 A0A1Z5KVB0 A0A2M4AN87 A0A1I8N0J4 T1PJB0 A0A1E1XBL9 A0A0L0CJW7 A0A1E1XL09 A0A131XXQ9 A0A1I8PVJ0 A0A1I8PVM2 W5L350 A0A226F2K3 V5GZ85 A0A1A9WDI6 A0A182H370 A0A1S3IL77 A0A3B4DRX5 A0A1D2NIX9 A0A210QAJ3 A0A224YKU9 A0A131YCY1 Q16HT7 A0A1S4G0D1 L7M0X7 A0A131YW63 W5JU93 A0A182W2G0 U4UHS3 A0A1B0AD56 A0A182K617 N6TYS3
Pubmed
28756777
26354079
22118469
18362917
19820115
23537049
+ More
28004739 24845553 26334808 25401762 27129103 24508170 26823975 30249741 20798317 21282665 21347285 21719571 28648823 20075255 20566863 25315136 22992520 28528879 28503490 26108605 29209593 25329095 25765539 26483478 27289101 28812685 28797301 26830274 17510324 25576852 20920257 23761445
28004739 24845553 26334808 25401762 27129103 24508170 26823975 30249741 20798317 21282665 21347285 21719571 28648823 20075255 20566863 25315136 22992520 28528879 28503490 26108605 29209593 25329095 25765539 26483478 27289101 28812685 28797301 26830274 17510324 25576852 20920257 23761445
EMBL
ODYU01000126
SOQ34381.1
KZ150152
PZC72826.1
NWSH01003436
PCG66309.1
+ More
KQ459053 KPJ04238.1 AGBW02008371 OWR53560.1 KQ460522 KPJ14161.1 GALX01000491 JAB67975.1 KQ971354 EFA06807.1 KB632322 ERL92356.1 APGK01058321 KB741288 ENN70657.1 GEZM01084002 JAV60860.1 GEDC01030990 JAS06308.1 NEVH01021939 PNF19124.1 PNF19114.1 PNF19127.1 KK855916 PTY24354.1 ACPB03008885 GEBQ01024578 JAT15399.1 KK852854 KDR14968.1 GECL01001395 JAP04729.1 GBGD01001126 JAC87763.1 GFTR01006541 JAW09885.1 GBRD01018159 JAG47668.1 GBHO01027776 JAG15828.1 GDKW01002536 JAI54059.1 KK107648 EZA48337.1 GDHC01021275 JAP97353.1 GDHC01010604 JAQ08025.1 QOIP01000008 RLU19353.1 GECU01024328 GECU01015307 JAS83378.1 JAS92399.1 GL438356 EFN69110.1 KQ434878 KZC09858.1 GDRN01070565 JAI63848.1 LBMM01005134 KMQ91790.1 GDRN01070560 JAI63852.1 GDRN01070561 JAI63851.1 GL762576 EFZ21088.1 KQ976394 KYM93318.1 ADTU01022032 GL450151 EFN81441.1 KQ980167 KYN17366.1 KQ769818 OAD52814.1 KQ981891 KYN33793.1 GL887974 EGI69336.1 GBYB01008149 JAG77916.1 NNAY01000154 OXU30499.1 KQ977120 KYN05562.1 KZ288416 PBC26053.1 KQ982422 KYQ56619.1 DS235818 EEB17666.1 JH431734 AJVK01005130 GGLE01000264 MBY04390.1 GEBQ01009501 JAT30476.1 GFWV01008996 MAA33725.1 JH815744 EKC27088.1 GGFK01008890 MBW42211.1 GFJQ02007960 JAV99009.1 GGFK01008923 MBW42244.1 KA648887 AFP63516.1 GFAC01002548 JAT96640.1 JRES01000303 KNC32507.1 GFAA01003540 JAT99894.1 GEFM01004791 JAP71005.1 LNIX01000001 OXA63708.1 GANP01008548 JAB75920.1 JXUM01106765 KQ565082 KXJ71364.1 LJIJ01000027 ODN05233.1 NEDP02004406 OWF45748.1 GFPF01007110 MAA18256.1 GEDV01012621 JAP75936.1 CH478142 EAT33817.1 GACK01008246 JAA56788.1 GEDV01006151 JAP82406.1 ADMH02000120 ETN67711.1 KB632169 ERL89425.1 APGK01049518 APGK01049519 KB741156 ENN73506.1
KQ459053 KPJ04238.1 AGBW02008371 OWR53560.1 KQ460522 KPJ14161.1 GALX01000491 JAB67975.1 KQ971354 EFA06807.1 KB632322 ERL92356.1 APGK01058321 KB741288 ENN70657.1 GEZM01084002 JAV60860.1 GEDC01030990 JAS06308.1 NEVH01021939 PNF19124.1 PNF19114.1 PNF19127.1 KK855916 PTY24354.1 ACPB03008885 GEBQ01024578 JAT15399.1 KK852854 KDR14968.1 GECL01001395 JAP04729.1 GBGD01001126 JAC87763.1 GFTR01006541 JAW09885.1 GBRD01018159 JAG47668.1 GBHO01027776 JAG15828.1 GDKW01002536 JAI54059.1 KK107648 EZA48337.1 GDHC01021275 JAP97353.1 GDHC01010604 JAQ08025.1 QOIP01000008 RLU19353.1 GECU01024328 GECU01015307 JAS83378.1 JAS92399.1 GL438356 EFN69110.1 KQ434878 KZC09858.1 GDRN01070565 JAI63848.1 LBMM01005134 KMQ91790.1 GDRN01070560 JAI63852.1 GDRN01070561 JAI63851.1 GL762576 EFZ21088.1 KQ976394 KYM93318.1 ADTU01022032 GL450151 EFN81441.1 KQ980167 KYN17366.1 KQ769818 OAD52814.1 KQ981891 KYN33793.1 GL887974 EGI69336.1 GBYB01008149 JAG77916.1 NNAY01000154 OXU30499.1 KQ977120 KYN05562.1 KZ288416 PBC26053.1 KQ982422 KYQ56619.1 DS235818 EEB17666.1 JH431734 AJVK01005130 GGLE01000264 MBY04390.1 GEBQ01009501 JAT30476.1 GFWV01008996 MAA33725.1 JH815744 EKC27088.1 GGFK01008890 MBW42211.1 GFJQ02007960 JAV99009.1 GGFK01008923 MBW42244.1 KA648887 AFP63516.1 GFAC01002548 JAT96640.1 JRES01000303 KNC32507.1 GFAA01003540 JAT99894.1 GEFM01004791 JAP71005.1 LNIX01000001 OXA63708.1 GANP01008548 JAB75920.1 JXUM01106765 KQ565082 KXJ71364.1 LJIJ01000027 ODN05233.1 NEDP02004406 OWF45748.1 GFPF01007110 MAA18256.1 GEDV01012621 JAP75936.1 CH478142 EAT33817.1 GACK01008246 JAA56788.1 GEDV01006151 JAP82406.1 ADMH02000120 ETN67711.1 KB632169 ERL89425.1 APGK01049518 APGK01049519 KB741156 ENN73506.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000007266
UP000030742
+ More
UP000019118 UP000192223 UP000235965 UP000015103 UP000027135 UP000053097 UP000279307 UP000000311 UP000076502 UP000036403 UP000078540 UP000005205 UP000008237 UP000078492 UP000078541 UP000007755 UP000215335 UP000078542 UP000242457 UP000075809 UP000002358 UP000005203 UP000009046 UP000092462 UP000075880 UP000095301 UP000005408 UP000037069 UP000095300 UP000018467 UP000198287 UP000091820 UP000069940 UP000249989 UP000085678 UP000261440 UP000094527 UP000242188 UP000008820 UP000000673 UP000075920 UP000092445 UP000075881
UP000019118 UP000192223 UP000235965 UP000015103 UP000027135 UP000053097 UP000279307 UP000000311 UP000076502 UP000036403 UP000078540 UP000005205 UP000008237 UP000078492 UP000078541 UP000007755 UP000215335 UP000078542 UP000242457 UP000075809 UP000002358 UP000005203 UP000009046 UP000092462 UP000075880 UP000095301 UP000005408 UP000037069 UP000095300 UP000018467 UP000198287 UP000091820 UP000069940 UP000249989 UP000085678 UP000261440 UP000094527 UP000242188 UP000008820 UP000000673 UP000075920 UP000092445 UP000075881
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
A0A2H1V0P3
A0A2W1BJ11
A0A2A4J360
A0A194QFB7
A0A212FIJ7
A0A0N0PCP1
+ More
V5GCW3 D6WPG0 U4UQG0 N6TWI2 A0A1Y1KKS6 A0A1W4XHY4 A0A1B6BYH9 A0A2J7PS11 A0A2J7PS04 A0A2J7PS14 A0A2R7WXT7 T1HMT7 A0A1B6KVC4 A0A067R6H4 A0A0V0G9Y9 A0A069DUF9 A0A224XBV9 A0A0K8S371 A0A0A9XAH5 A0A0P4VNY7 A0A026VX52 A0A146KLC0 A0A146LLA5 A0A3L8DGQ1 A0A1B6I8Y1 E2ABT3 A0A154PEY2 A0A0N7ZC98 A0A0J7NGY9 A0A0P4W5J3 A0A0P4WC21 E9IED2 A0A195BZH9 A0A158NP13 E2BS49 A0A195DX02 A0A310S5J5 A0A195EZZ2 F4W8X7 A0A0C9QWN8 A0A232FJ39 A0A151ILD2 A0A2A3E2T3 A0A151X8F6 K7J170 A0A088ADV5 E0VWB0 T1J077 A0A1B0DCJ2 A0A2R5L4M2 A0A182IW73 A0A1B6M3G4 A0A1I8N0H4 A0A293M422 K1PZM5 A0A2M4AN58 A0A1Z5KVB0 A0A2M4AN87 A0A1I8N0J4 T1PJB0 A0A1E1XBL9 A0A0L0CJW7 A0A1E1XL09 A0A131XXQ9 A0A1I8PVJ0 A0A1I8PVM2 W5L350 A0A226F2K3 V5GZ85 A0A1A9WDI6 A0A182H370 A0A1S3IL77 A0A3B4DRX5 A0A1D2NIX9 A0A210QAJ3 A0A224YKU9 A0A131YCY1 Q16HT7 A0A1S4G0D1 L7M0X7 A0A131YW63 W5JU93 A0A182W2G0 U4UHS3 A0A1B0AD56 A0A182K617 N6TYS3
V5GCW3 D6WPG0 U4UQG0 N6TWI2 A0A1Y1KKS6 A0A1W4XHY4 A0A1B6BYH9 A0A2J7PS11 A0A2J7PS04 A0A2J7PS14 A0A2R7WXT7 T1HMT7 A0A1B6KVC4 A0A067R6H4 A0A0V0G9Y9 A0A069DUF9 A0A224XBV9 A0A0K8S371 A0A0A9XAH5 A0A0P4VNY7 A0A026VX52 A0A146KLC0 A0A146LLA5 A0A3L8DGQ1 A0A1B6I8Y1 E2ABT3 A0A154PEY2 A0A0N7ZC98 A0A0J7NGY9 A0A0P4W5J3 A0A0P4WC21 E9IED2 A0A195BZH9 A0A158NP13 E2BS49 A0A195DX02 A0A310S5J5 A0A195EZZ2 F4W8X7 A0A0C9QWN8 A0A232FJ39 A0A151ILD2 A0A2A3E2T3 A0A151X8F6 K7J170 A0A088ADV5 E0VWB0 T1J077 A0A1B0DCJ2 A0A2R5L4M2 A0A182IW73 A0A1B6M3G4 A0A1I8N0H4 A0A293M422 K1PZM5 A0A2M4AN58 A0A1Z5KVB0 A0A2M4AN87 A0A1I8N0J4 T1PJB0 A0A1E1XBL9 A0A0L0CJW7 A0A1E1XL09 A0A131XXQ9 A0A1I8PVJ0 A0A1I8PVM2 W5L350 A0A226F2K3 V5GZ85 A0A1A9WDI6 A0A182H370 A0A1S3IL77 A0A3B4DRX5 A0A1D2NIX9 A0A210QAJ3 A0A224YKU9 A0A131YCY1 Q16HT7 A0A1S4G0D1 L7M0X7 A0A131YW63 W5JU93 A0A182W2G0 U4UHS3 A0A1B0AD56 A0A182K617 N6TYS3
PDB
3NF1
E-value=0.000220853,
Score=107
Ontologies
GO
PANTHER
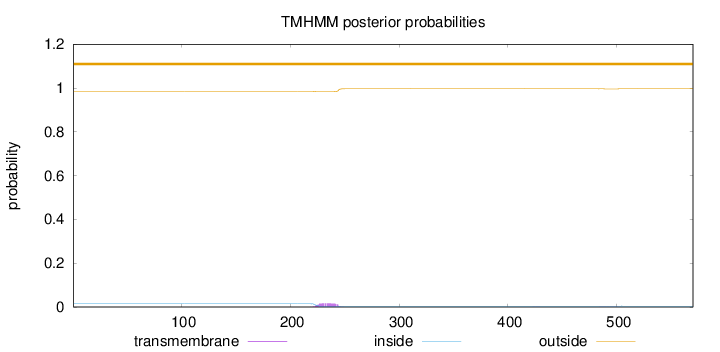
Topology
Length:
570
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.33575
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01485
outside
1 - 570
Population Genetic Test Statistics
Pi
218.664683
Theta
138.308615
Tajima's D
1.495246
CLR
0.260751
CSRT
0.780910954452277
Interpretation
Uncertain
