Gene
KWMTBOMO12235
Annotation
PREDICTED:_uncharacterized_protein_LOC106139904_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.753
Sequence
CDS
ATGCATACCTGTGCGAATTGTGGTACTCAACACACCGACGGAACAATATGCAGCATTTGCAAGAATCATTATGATTTTCCATGTTCTGGAGTGACGGAATCCGGCTATCGCAGATTAGCAACAGTGGTGGAAGACATTAAGAGCATTAAGGCTGAAATAGGCGATCTCAAGAATTCCCTCGAGCTGGCATATCAACTCACTGGTGATCTCTCGAACACGGTGAAGGACTTGGGCACTAGGATATGTACAGTTGAAAATATCGGCAGTGAGGTTCCTGCTCTTCGTGACGAAATTACCAGGTTGAACAATGAGCTACAGGACCGGGACCAGTGGGCCCGTGCGAATAACGTGGAGATCCGTGGTATACCGGAAAAGAAGGGTGAAAATCTATATGATGTCGTCAGCTGTATCGGCCAACTTTATAATATTTCCATTACGAAAGAGGAGATTAATTACATTGCACGTATCCCAACTCGTGTGCTGAAGGCTGAAAAATCTATTATTGTTGCGTTCAACAATAGGTATCGCAAGGAGGAACTTATCGCTTTAGCTCGTAAAATTAAGAAGAACAGTCTCTCGGACTTGGGTATACGAGGAGAGGGAAAATTTTATCTGAACGACCATTTGACGCAGCGTAATAAAACATTATTAAACAAAGCAAAGTCGCTCGCGAAGGATAATAATTTCCAATATATTTGGGTAAAACATTGTAAAATAATGGCCCGCAAATCAGATACATCCCCGATATTTTTTATAAAAGAGGAAAGGGATTTGAAGAAGATAACCTGA
Protein
MHTCANCGTQHTDGTICSICKNHYDFPCSGVTESGYRRLATVVEDIKSIKAEIGDLKNSLELAYQLTGDLSNTVKDLGTRICTVENIGSEVPALRDEITRLNNELQDRDQWARANNVEIRGIPEKKGENLYDVVSCIGQLYNISITKEEINYIARIPTRVLKAEKSIIVAFNNRYRKEELIALARKIKKNSLSDLGIRGEGKFYLNDHLTQRNKTLLNKAKSLAKDNNFQYIWVKHCKIMARKSDTSPIFFIKEERDLKKIT
Summary
Uniprot
H9JUF4
A0A2A4K238
A0A0L7KMA6
A0A2W1BT79
A0A2H1WI81
A0A0L7K2C3
+ More
A0A0L7LF31 H9J7H5 A0A0L7LM62 A0A2A4JLA2 A0A194PTX1 A0A2W1BZ95 A0A2A4JCE9 A0A2H1W809 A0A0L7K4U1 A0A0L7KYR8 A0A0L7K2T7 A0A2A4IYL5 A0A0L7L6I7 A0A2A4JUE7 A0A2H1WBK3 A0A0L7LIM3 A0A0L7LR61 A0A0L7L7A2 A0A212F6C5 A0A0L7LF48 A0A0L7K2E3 A0A1E1WAX3 A0A2A4J140 A0A0L7LQ84 A0A2A4JSX1 A0A0L7LS47 A0A0L7LEI8 G3E4N3 A0A0L7L8J7 A0A2A4IYM6 A0A2A4JR78 A0A0L7KMZ2 A0A194QTI2 A0A0A9WQ01 A0A1Y1KQY7 A0A1S3DD01 A0A1E1WIH3 A0A2A4JD31 A0A1Y1LTU7 A0A1Y1L521 A0A2A4J6R0 A0A293LR31 A0A1E1X2K3 A0A2H1X1K9 A0A2A4IZ17 A0A2W1BEV5 A0A2A4ITG1 A0A0L7L118 A0A0L7LCM2 A0A2A4J6Z0 A0A2A4JWA7 A0A1S3DCA1 A0A1S3DH78 A0A2W1C270 A0A2H1WNV1 A0A0C9S1R0 A0A2H1VKR1 A0A1B6LMZ9 A0A0L7KTU7 A0A2A4J6A9 A0A2H1WRH7 A0A1B6K2E9 A0A2H1WIH0 A0A1B6ITU2 A0A2A4J8T1 A0A0L7L1J8 A0A0L7LEU2 A0A0L7KR27 A0A0L7LR99 A0A1Y1LUM5 J9JYP6
A0A0L7LF31 H9J7H5 A0A0L7LM62 A0A2A4JLA2 A0A194PTX1 A0A2W1BZ95 A0A2A4JCE9 A0A2H1W809 A0A0L7K4U1 A0A0L7KYR8 A0A0L7K2T7 A0A2A4IYL5 A0A0L7L6I7 A0A2A4JUE7 A0A2H1WBK3 A0A0L7LIM3 A0A0L7LR61 A0A0L7L7A2 A0A212F6C5 A0A0L7LF48 A0A0L7K2E3 A0A1E1WAX3 A0A2A4J140 A0A0L7LQ84 A0A2A4JSX1 A0A0L7LS47 A0A0L7LEI8 G3E4N3 A0A0L7L8J7 A0A2A4IYM6 A0A2A4JR78 A0A0L7KMZ2 A0A194QTI2 A0A0A9WQ01 A0A1Y1KQY7 A0A1S3DD01 A0A1E1WIH3 A0A2A4JD31 A0A1Y1LTU7 A0A1Y1L521 A0A2A4J6R0 A0A293LR31 A0A1E1X2K3 A0A2H1X1K9 A0A2A4IZ17 A0A2W1BEV5 A0A2A4ITG1 A0A0L7L118 A0A0L7LCM2 A0A2A4J6Z0 A0A2A4JWA7 A0A1S3DCA1 A0A1S3DH78 A0A2W1C270 A0A2H1WNV1 A0A0C9S1R0 A0A2H1VKR1 A0A1B6LMZ9 A0A0L7KTU7 A0A2A4J6A9 A0A2H1WRH7 A0A1B6K2E9 A0A2H1WIH0 A0A1B6ITU2 A0A2A4J8T1 A0A0L7L1J8 A0A0L7LEU2 A0A0L7KR27 A0A0L7LR99 A0A1Y1LUM5 J9JYP6
Pubmed
EMBL
BABH01022797
BABH01022798
BABH01022799
BABH01022800
BABH01022801
BABH01022802
+ More
NWSH01000216 PCG78307.1 JTDY01009133 KOB64064.1 KZ150018 PZC74933.1 ODYU01008832 SOQ52780.1 JTDY01017795 JTDY01002629 KOB51833.1 KOB71066.1 JTDY01001364 KOB74072.1 BABH01013843 JTDY01000621 KOB76444.1 NWSH01001181 PCG72223.1 KQ459592 KPI96861.1 KZ149893 PZC78914.1 NWSH01001977 PCG69519.1 ODYU01006867 SOQ49096.1 JTDY01010706 KOB58083.1 JTDY01004228 KOB68423.1 JTDY01013395 KOB52145.1 NWSH01004950 PCG64556.1 KOB71067.1 NWSH01000638 PCG75103.1 ODYU01007576 SOQ50461.1 JTDY01000967 KOB75255.1 JTDY01000335 KOB77651.1 JTDY01006475 JTDY01002579 KOB65982.1 KOB71161.1 AGBW02010051 OWR49283.1 JTDY01001455 KOB73826.1 JTDY01015045 KOB51976.1 GDQN01006959 JAT84095.1 NWSH01003729 PCG65877.1 JTDY01000399 KOB77361.1 NWSH01000633 PCG75137.1 JTDY01000225 KOB78204.1 JTDY01001427 KOB73882.1 HQ585015 AEM44816.1 JTDY01002226 KOB71853.1 NWSH01004492 PCG65047.1 NWSH01000830 PCG73963.1 JTDY01008292 KOB64643.1 KQ461154 KPJ08619.1 GBHO01033077 JAG10527.1 GEZM01076104 JAV63822.1 GDQN01004260 JAT86794.1 NWSH01002051 PCG69303.1 GEZM01051380 GEZM01051375 JAV74996.1 GEZM01064388 JAV68789.1 NWSH01002710 PCG67641.1 GFWV01006244 MAA30974.1 GFAC01005699 JAT93489.1 ODYU01012701 SOQ59142.1 NWSH01005158 PCG64392.1 KZ150150 PZC72841.1 NWSH01008646 PCG62462.1 JTDY01003781 KOB69016.1 JTDY01001653 KOB73242.1 NWSH01002711 PCG67639.1 NWSH01000452 NWSH01000043 PCG76317.1 PCG80309.1 KZ149908 PZC78213.1 ODYU01009977 SOQ54751.1 GBZX01001916 JAG90824.1 ODYU01002850 SOQ40844.1 GEBQ01014872 JAT25105.1 JTDY01005854 KOB66545.1 NWSH01002730 PCG67615.1 ODYU01010507 SOQ55661.1 GECU01002092 JAT05615.1 ODYU01008890 SOQ52873.1 GECU01017357 JAS90349.1 NWSH01002492 PCG68176.1 JTDY01003535 KOB69383.1 JTDY01001361 KOB74078.1 JTDY01007036 KOB65506.1 JTDY01000273 KOB77970.1 GEZM01048403 JAV76588.1 ABLF02018094 ABLF02018096
NWSH01000216 PCG78307.1 JTDY01009133 KOB64064.1 KZ150018 PZC74933.1 ODYU01008832 SOQ52780.1 JTDY01017795 JTDY01002629 KOB51833.1 KOB71066.1 JTDY01001364 KOB74072.1 BABH01013843 JTDY01000621 KOB76444.1 NWSH01001181 PCG72223.1 KQ459592 KPI96861.1 KZ149893 PZC78914.1 NWSH01001977 PCG69519.1 ODYU01006867 SOQ49096.1 JTDY01010706 KOB58083.1 JTDY01004228 KOB68423.1 JTDY01013395 KOB52145.1 NWSH01004950 PCG64556.1 KOB71067.1 NWSH01000638 PCG75103.1 ODYU01007576 SOQ50461.1 JTDY01000967 KOB75255.1 JTDY01000335 KOB77651.1 JTDY01006475 JTDY01002579 KOB65982.1 KOB71161.1 AGBW02010051 OWR49283.1 JTDY01001455 KOB73826.1 JTDY01015045 KOB51976.1 GDQN01006959 JAT84095.1 NWSH01003729 PCG65877.1 JTDY01000399 KOB77361.1 NWSH01000633 PCG75137.1 JTDY01000225 KOB78204.1 JTDY01001427 KOB73882.1 HQ585015 AEM44816.1 JTDY01002226 KOB71853.1 NWSH01004492 PCG65047.1 NWSH01000830 PCG73963.1 JTDY01008292 KOB64643.1 KQ461154 KPJ08619.1 GBHO01033077 JAG10527.1 GEZM01076104 JAV63822.1 GDQN01004260 JAT86794.1 NWSH01002051 PCG69303.1 GEZM01051380 GEZM01051375 JAV74996.1 GEZM01064388 JAV68789.1 NWSH01002710 PCG67641.1 GFWV01006244 MAA30974.1 GFAC01005699 JAT93489.1 ODYU01012701 SOQ59142.1 NWSH01005158 PCG64392.1 KZ150150 PZC72841.1 NWSH01008646 PCG62462.1 JTDY01003781 KOB69016.1 JTDY01001653 KOB73242.1 NWSH01002711 PCG67639.1 NWSH01000452 NWSH01000043 PCG76317.1 PCG80309.1 KZ149908 PZC78213.1 ODYU01009977 SOQ54751.1 GBZX01001916 JAG90824.1 ODYU01002850 SOQ40844.1 GEBQ01014872 JAT25105.1 JTDY01005854 KOB66545.1 NWSH01002730 PCG67615.1 ODYU01010507 SOQ55661.1 GECU01002092 JAT05615.1 ODYU01008890 SOQ52873.1 GECU01017357 JAS90349.1 NWSH01002492 PCG68176.1 JTDY01003535 KOB69383.1 JTDY01001361 KOB74078.1 JTDY01007036 KOB65506.1 JTDY01000273 KOB77970.1 GEZM01048403 JAV76588.1 ABLF02018094 ABLF02018096
Proteomes
Interpro
IPR013083
Znf_RING/FYVE/PHD
+ More
IPR004244 Transposase_22
IPR004941 Baculo_FP
IPR011011 Znf_FYVE_PHD
IPR019787 Znf_PHD-finger
IPR019786 Zinc_finger_PHD-type_CS
IPR001965 Znf_PHD
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR007311 ST7
IPR001841 Znf_RING
IPR002219 PE/DAG-bd
IPR004244 Transposase_22
IPR004941 Baculo_FP
IPR011011 Znf_FYVE_PHD
IPR019787 Znf_PHD-finger
IPR019786 Zinc_finger_PHD-type_CS
IPR001965 Znf_PHD
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR007311 ST7
IPR001841 Znf_RING
IPR002219 PE/DAG-bd
Gene 3D
ProteinModelPortal
H9JUF4
A0A2A4K238
A0A0L7KMA6
A0A2W1BT79
A0A2H1WI81
A0A0L7K2C3
+ More
A0A0L7LF31 H9J7H5 A0A0L7LM62 A0A2A4JLA2 A0A194PTX1 A0A2W1BZ95 A0A2A4JCE9 A0A2H1W809 A0A0L7K4U1 A0A0L7KYR8 A0A0L7K2T7 A0A2A4IYL5 A0A0L7L6I7 A0A2A4JUE7 A0A2H1WBK3 A0A0L7LIM3 A0A0L7LR61 A0A0L7L7A2 A0A212F6C5 A0A0L7LF48 A0A0L7K2E3 A0A1E1WAX3 A0A2A4J140 A0A0L7LQ84 A0A2A4JSX1 A0A0L7LS47 A0A0L7LEI8 G3E4N3 A0A0L7L8J7 A0A2A4IYM6 A0A2A4JR78 A0A0L7KMZ2 A0A194QTI2 A0A0A9WQ01 A0A1Y1KQY7 A0A1S3DD01 A0A1E1WIH3 A0A2A4JD31 A0A1Y1LTU7 A0A1Y1L521 A0A2A4J6R0 A0A293LR31 A0A1E1X2K3 A0A2H1X1K9 A0A2A4IZ17 A0A2W1BEV5 A0A2A4ITG1 A0A0L7L118 A0A0L7LCM2 A0A2A4J6Z0 A0A2A4JWA7 A0A1S3DCA1 A0A1S3DH78 A0A2W1C270 A0A2H1WNV1 A0A0C9S1R0 A0A2H1VKR1 A0A1B6LMZ9 A0A0L7KTU7 A0A2A4J6A9 A0A2H1WRH7 A0A1B6K2E9 A0A2H1WIH0 A0A1B6ITU2 A0A2A4J8T1 A0A0L7L1J8 A0A0L7LEU2 A0A0L7KR27 A0A0L7LR99 A0A1Y1LUM5 J9JYP6
A0A0L7LF31 H9J7H5 A0A0L7LM62 A0A2A4JLA2 A0A194PTX1 A0A2W1BZ95 A0A2A4JCE9 A0A2H1W809 A0A0L7K4U1 A0A0L7KYR8 A0A0L7K2T7 A0A2A4IYL5 A0A0L7L6I7 A0A2A4JUE7 A0A2H1WBK3 A0A0L7LIM3 A0A0L7LR61 A0A0L7L7A2 A0A212F6C5 A0A0L7LF48 A0A0L7K2E3 A0A1E1WAX3 A0A2A4J140 A0A0L7LQ84 A0A2A4JSX1 A0A0L7LS47 A0A0L7LEI8 G3E4N3 A0A0L7L8J7 A0A2A4IYM6 A0A2A4JR78 A0A0L7KMZ2 A0A194QTI2 A0A0A9WQ01 A0A1Y1KQY7 A0A1S3DD01 A0A1E1WIH3 A0A2A4JD31 A0A1Y1LTU7 A0A1Y1L521 A0A2A4J6R0 A0A293LR31 A0A1E1X2K3 A0A2H1X1K9 A0A2A4IZ17 A0A2W1BEV5 A0A2A4ITG1 A0A0L7L118 A0A0L7LCM2 A0A2A4J6Z0 A0A2A4JWA7 A0A1S3DCA1 A0A1S3DH78 A0A2W1C270 A0A2H1WNV1 A0A0C9S1R0 A0A2H1VKR1 A0A1B6LMZ9 A0A0L7KTU7 A0A2A4J6A9 A0A2H1WRH7 A0A1B6K2E9 A0A2H1WIH0 A0A1B6ITU2 A0A2A4J8T1 A0A0L7L1J8 A0A0L7LEU2 A0A0L7KR27 A0A0L7LR99 A0A1Y1LUM5 J9JYP6
Ontologies
Topology
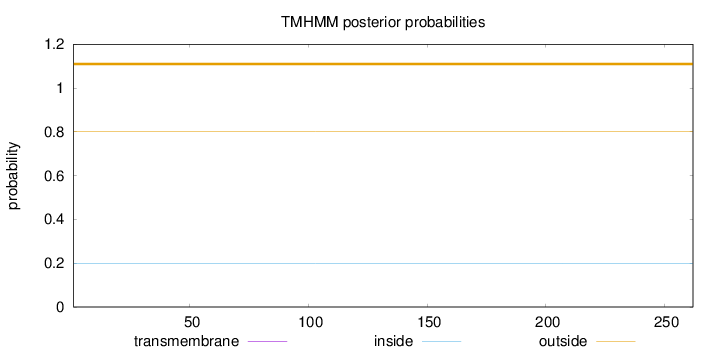
Length:
262
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00055
Exp number, first 60 AAs:
0
Total prob of N-in:
0.19962
outside
1 - 262
Population Genetic Test Statistics
Pi
203.21356
Theta
139.279787
Tajima's D
1.278439
CLR
1.363283
CSRT
0.733013349332533
Interpretation
Uncertain
