Gene
KWMTBOMO12233
Pre Gene Modal
BGIBMGA014191
Annotation
PREDICTED:_myrosinase_1-like_[Papilio_xuthus]
Location in the cell
Lysosomal Reliability : 1.938
Sequence
CDS
ATGCTTGTAGAACTAGGCGTACACTTTTATAGGTTCTCAATATCCTGGCCACGAGTTCTGCCCACCGGTTTTCCAAACTACATCAGCGAAGATGGAGTTAAATATTATAATACTTTGATAAATGCGTTGTTGGAAAACGGAATAGAGCCCTTGGTTACTTTGTACCATTGGGACCTTCCACAGAGACTCCAGGACTTAGGAGGCTGGACGAATCCTCACATCGCAGAATGGTTCGAAAATTACGCCAGAACAGCCTTCGAGTTATTTGGCGACAGAGTGAAGACTTGGATCACGATTAATGAGCCTTTCGTCGTTTGCGACATGGCGTACAACACTGGTCTGGTGGCTCCTGGGATCATTGATCCTGAAATTGCACCATATCTGTGCAATAAATACACGTTGATGGCCCACGCGAGAGCCTGGAGACTTTACGATAAGGAGTTCAGACATAAATATCATGGTAAAATCTCCATCGCGAACCAATTGTTTTGGTTTCAGCCTCTTACTGATGAAGATAAAGAAGTCACTGAACTAGCATTGGAGAATTGTGTCGGTCGGTACTCTCATCCAATTTACTCTAAGAAAGGTGGATGGCCGCCGAGCATAGAGAAAACTATCCATGACAATAGTCGAAGACAAGGGTACCTGAAGTCAAATTTACCCGTTCTGACTAAGCAAGAGATTGAGTACATCAAGGGTACCTATGACTTCTTTGGACTGAATCACTACACAAGTCGCTTGGTCCGAAAAGCCGAGACTGGCGAGAAGCTGAGTCCCTGGCCCATAGGGGATGCTCCTGATTTCGATGCTCGACTTCTAGTTTCGCCGCACTGGCCCAAGACTTCATCTTCTTGGTTTTTTGTCAACCCACCAGGGATAAGAAAACTGCTGCAGTGGTTGAAGAAAAATTACGGCGATCTTACAGTTTTAGTAACGGAAAACGGATTTCCGAATCTTGGTGGCGTGGAGGATGTTGACCGAATAAACTATTACAGAGATAATATGCATGAGATTCTCCTGGCTATCAAAAAAGACAAAATCAACGTAACCGCATACACGGCTTGGACGCTGATGGACAACTACGAATGGATGGATGGATACAAGTGA
Protein
MLVELGVHFYRFSISWPRVLPTGFPNYISEDGVKYYNTLINALLENGIEPLVTLYHWDLPQRLQDLGGWTNPHIAEWFENYARTAFELFGDRVKTWITINEPFVVCDMAYNTGLVAPGIIDPEIAPYLCNKYTLMAHARAWRLYDKEFRHKYHGKISIANQLFWFQPLTDEDKEVTELALENCVGRYSHPIYSKKGGWPPSIEKTIHDNSRRQGYLKSNLPVLTKQEIEYIKGTYDFFGLNHYTSRLVRKAETGEKLSPWPIGDAPDFDARLLVSPHWPKTSSSWFFVNPPGIRKLLQWLKKNYGDLTVLVTENGFPNLGGVEDVDRINYYRDNMHEILLAIKKDKINVTAYTAWTLMDNYEWMDGYK
Summary
Similarity
Belongs to the glycosyl hydrolase 1 family.
Belongs to the class-I aminoacyl-tRNA synthetase family.
Belongs to the class-I aminoacyl-tRNA synthetase family.
Uniprot
H9JXC6
A0A2A4J905
A0A2W1BMB0
A0A2H1VC49
A0A2H1W6N8
A0A2W1BC39
+ More
A0A2A4IWH6 A0A194QH27 A0A194QFH9 H9JXC7 A0A3G1T1E9 A0A194QFC2 H9JX79 H9JXC5 A0A3S2N597 A0A2W1BEU6 A0A2A4IZ96 G9F9G6 A0A2H1WLJ0 A0A2H1WLB6 A0A194QLC3 A0A2W1BJB1 A0A2W1BD53 A0A0N1I6T9 H9JX83 A0A0N1IBS6 A0A212FIL5 A0A2H1VX30 A0A194R3H3 A0A194PPG4 A0A1Q3FR87 A0A2H1V873 A0A1B6E1F2 A0A1B6D5W8 A0A0T6B9C6 B0WNN7 A0A2A4JEG9 A0A3S2PE71 Q5TQY8 A0A182U0B4 A0A1Y9IRY8 D0VYS1 A0A1Y9GKT0 A0A1Y9J100 G9F9I3 A0A2H1WZM4 D5KQK3 H9JX80 A0A1Y1K2R8 H9IZH5 A0A1S4FLR9 A0A182GJ50 A0A212F963 A0A212EKQ3 D0VYR5 A0A194PMG8 D0VYR6 G9F9G9 D0VYR8 Q16WF2 A0A182QI13 D0VYS0 A0A1S4G3W5 A0A2M3ZET6 A0A194PVV9 A0A1Y9IW73 A0A2M4BKN2 Q16ET6 W5JN46 H9JXC4 A0A212EXF2 A0A212FF93 A0A194R3H6 A0A212FF90 A0A2J7PTJ0 A0A182HAK7 Q8T0W7 S5SP57 A0A2W1BNB0 A0A182J417 A0A182LL23 D2X5V7 A0A3S2TR57 A0A194R360 A0A194QDS5 A0A0N1PFL9 D0VYR9 H9JLY4 A0A194PMT2 A0A2H1VWC6 G9F9I2 D9ILU1 A0A1B0CB38 D6X3T6 A0A1Y1KBB7 A0A182YP84 A0A212FBG8 H9JLY5 A0A2J7PKH6 A0A165G9G6
A0A2A4IWH6 A0A194QH27 A0A194QFH9 H9JXC7 A0A3G1T1E9 A0A194QFC2 H9JX79 H9JXC5 A0A3S2N597 A0A2W1BEU6 A0A2A4IZ96 G9F9G6 A0A2H1WLJ0 A0A2H1WLB6 A0A194QLC3 A0A2W1BJB1 A0A2W1BD53 A0A0N1I6T9 H9JX83 A0A0N1IBS6 A0A212FIL5 A0A2H1VX30 A0A194R3H3 A0A194PPG4 A0A1Q3FR87 A0A2H1V873 A0A1B6E1F2 A0A1B6D5W8 A0A0T6B9C6 B0WNN7 A0A2A4JEG9 A0A3S2PE71 Q5TQY8 A0A182U0B4 A0A1Y9IRY8 D0VYS1 A0A1Y9GKT0 A0A1Y9J100 G9F9I3 A0A2H1WZM4 D5KQK3 H9JX80 A0A1Y1K2R8 H9IZH5 A0A1S4FLR9 A0A182GJ50 A0A212F963 A0A212EKQ3 D0VYR5 A0A194PMG8 D0VYR6 G9F9G9 D0VYR8 Q16WF2 A0A182QI13 D0VYS0 A0A1S4G3W5 A0A2M3ZET6 A0A194PVV9 A0A1Y9IW73 A0A2M4BKN2 Q16ET6 W5JN46 H9JXC4 A0A212EXF2 A0A212FF93 A0A194R3H6 A0A212FF90 A0A2J7PTJ0 A0A182HAK7 Q8T0W7 S5SP57 A0A2W1BNB0 A0A182J417 A0A182LL23 D2X5V7 A0A3S2TR57 A0A194R360 A0A194QDS5 A0A0N1PFL9 D0VYR9 H9JLY4 A0A194PMT2 A0A2H1VWC6 G9F9I2 D9ILU1 A0A1B0CB38 D6X3T6 A0A1Y1KBB7 A0A182YP84 A0A212FBG8 H9JLY5 A0A2J7PKH6 A0A165G9G6
Pubmed
EMBL
BABH01035833
BABH01035834
BABH01035835
NWSH01002416
PCG68339.1
KZ150152
+ More
PZC72823.1 ODYU01001752 SOQ38385.1 ODYU01006661 SOQ48703.1 PZC72822.1 NWSH01005512 PCG64155.1 KQ459053 KPJ04240.1 KPJ04242.1 BABH01035838 BABH01035839 MG992371 AXY94809.1 KPJ04243.1 BABH01035825 BABH01035826 BABH01035827 RSAL01000405 RVE41892.1 PZC72831.1 NWSH01004417 PCG65145.1 JN033711 AEW46864.1 ODYU01009444 SOQ53877.1 SOQ53875.1 KPJ04241.1 PZC72820.1 KZ150122 PZC73162.1 KQ461157 KPJ08090.1 BABH01035811 BABH01035812 KPJ08092.1 AGBW02008370 OWR53561.1 ODYU01004975 SOQ45390.1 KQ460779 KPJ12242.1 KQ459598 KPI94624.1 GFDL01004884 JAV30161.1 ODYU01001184 SOQ37050.1 GEDC01005540 JAS31758.1 GEDC01016220 JAS21078.1 LJIG01003002 KRT83938.1 DS232013 EDS31873.1 NWSH01001829 PCG69984.1 RSAL01000077 RVE48784.1 AAAB01008960 EAL40075.2 AB508960 BAI50024.1 APCN01002262 JN033728 AEW46881.1 ODYU01012291 SOQ58525.1 GU591172 ADD92156.1 BABH01035816 BABH01035817 GEZM01098374 GEZM01098373 JAV53905.1 BABH01014157 JXUM01012550 JXUM01012551 JXUM01012552 KQ560357 KXJ82853.1 AGBW02009638 OWR50268.1 AGBW02014221 OWR42063.1 AB508954 AB508956 BAI50018.1 BAI50020.1 KPI94631.1 AB508955 BAI50019.1 JN033714 AEW46867.1 AB508957 BAI50021.1 CH477568 EAT38910.1 AXCN02000455 AB508959 BAI50023.1 GGFM01006254 MBW27005.1 KQ459589 KPI97521.1 GGFJ01004382 MBW53523.1 CH478582 EAT32750.1 ADMH02000892 ETN64738.1 BABH01035821 AGBW02011758 OWR46178.1 AGBW02008847 OWR52397.1 KPJ12247.1 OWR52398.1 NEVH01021237 PNF19653.1 JXUM01031352 KQ560919 KXJ80348.1 AB073638 BAB91145.1 KC891004 AGS32242.1 PZC73163.1 GQ911585 ADB23476.1 RSAL01000018 RVE52880.1 KPJ12243.1 KQ459144 KPJ03612.1 KPJ08091.1 AB508958 BAI50022.1 BABH01005437 KPI94627.1 ODYU01004836 SOQ45125.1 JN033727 AEW46880.1 HM152540 ADK12988.1 AJWK01004788 AJWK01004789 AJWK01004790 KQ971326 EEZ97373.2 GEZM01089030 GEZM01089029 JAV57758.1 AGBW02009344 OWR51063.1 BABH01005438 NEVH01024540 PNF16838.1 KU886065 AMY26906.1
PZC72823.1 ODYU01001752 SOQ38385.1 ODYU01006661 SOQ48703.1 PZC72822.1 NWSH01005512 PCG64155.1 KQ459053 KPJ04240.1 KPJ04242.1 BABH01035838 BABH01035839 MG992371 AXY94809.1 KPJ04243.1 BABH01035825 BABH01035826 BABH01035827 RSAL01000405 RVE41892.1 PZC72831.1 NWSH01004417 PCG65145.1 JN033711 AEW46864.1 ODYU01009444 SOQ53877.1 SOQ53875.1 KPJ04241.1 PZC72820.1 KZ150122 PZC73162.1 KQ461157 KPJ08090.1 BABH01035811 BABH01035812 KPJ08092.1 AGBW02008370 OWR53561.1 ODYU01004975 SOQ45390.1 KQ460779 KPJ12242.1 KQ459598 KPI94624.1 GFDL01004884 JAV30161.1 ODYU01001184 SOQ37050.1 GEDC01005540 JAS31758.1 GEDC01016220 JAS21078.1 LJIG01003002 KRT83938.1 DS232013 EDS31873.1 NWSH01001829 PCG69984.1 RSAL01000077 RVE48784.1 AAAB01008960 EAL40075.2 AB508960 BAI50024.1 APCN01002262 JN033728 AEW46881.1 ODYU01012291 SOQ58525.1 GU591172 ADD92156.1 BABH01035816 BABH01035817 GEZM01098374 GEZM01098373 JAV53905.1 BABH01014157 JXUM01012550 JXUM01012551 JXUM01012552 KQ560357 KXJ82853.1 AGBW02009638 OWR50268.1 AGBW02014221 OWR42063.1 AB508954 AB508956 BAI50018.1 BAI50020.1 KPI94631.1 AB508955 BAI50019.1 JN033714 AEW46867.1 AB508957 BAI50021.1 CH477568 EAT38910.1 AXCN02000455 AB508959 BAI50023.1 GGFM01006254 MBW27005.1 KQ459589 KPI97521.1 GGFJ01004382 MBW53523.1 CH478582 EAT32750.1 ADMH02000892 ETN64738.1 BABH01035821 AGBW02011758 OWR46178.1 AGBW02008847 OWR52397.1 KPJ12247.1 OWR52398.1 NEVH01021237 PNF19653.1 JXUM01031352 KQ560919 KXJ80348.1 AB073638 BAB91145.1 KC891004 AGS32242.1 PZC73163.1 GQ911585 ADB23476.1 RSAL01000018 RVE52880.1 KPJ12243.1 KQ459144 KPJ03612.1 KPJ08091.1 AB508958 BAI50022.1 BABH01005437 KPI94627.1 ODYU01004836 SOQ45125.1 JN033727 AEW46880.1 HM152540 ADK12988.1 AJWK01004788 AJWK01004789 AJWK01004790 KQ971326 EEZ97373.2 GEZM01089030 GEZM01089029 JAV57758.1 AGBW02009344 OWR51063.1 BABH01005438 NEVH01024540 PNF16838.1 KU886065 AMY26906.1
Proteomes
Pfam
Interpro
IPR033132
Glyco_hydro_1_N_CS
+ More
IPR001360 Glyco_hydro_1
IPR017853 Glycoside_hydrolase_SF
IPR018120 Glyco_hydro_1_AS
IPR000873 AMP-dep_Synth/Lig
IPR025110 AMP-bd_C
IPR002300 aa-tRNA-synth_Ia
IPR014729 Rossmann-like_a/b/a_fold
IPR001412 aa-tRNA-synth_I_CS
IPR009008 Val/Leu/Ile-tRNA-synth_edit
IPR002301 Ile-tRNA-ligase
IPR011234 Fumarylacetoacetase-like_C
IPR036663 Fumarylacetoacetase_C_sf
IPR001360 Glyco_hydro_1
IPR017853 Glycoside_hydrolase_SF
IPR018120 Glyco_hydro_1_AS
IPR000873 AMP-dep_Synth/Lig
IPR025110 AMP-bd_C
IPR002300 aa-tRNA-synth_Ia
IPR014729 Rossmann-like_a/b/a_fold
IPR001412 aa-tRNA-synth_I_CS
IPR009008 Val/Leu/Ile-tRNA-synth_edit
IPR002301 Ile-tRNA-ligase
IPR011234 Fumarylacetoacetase-like_C
IPR036663 Fumarylacetoacetase_C_sf
Gene 3D
ProteinModelPortal
H9JXC6
A0A2A4J905
A0A2W1BMB0
A0A2H1VC49
A0A2H1W6N8
A0A2W1BC39
+ More
A0A2A4IWH6 A0A194QH27 A0A194QFH9 H9JXC7 A0A3G1T1E9 A0A194QFC2 H9JX79 H9JXC5 A0A3S2N597 A0A2W1BEU6 A0A2A4IZ96 G9F9G6 A0A2H1WLJ0 A0A2H1WLB6 A0A194QLC3 A0A2W1BJB1 A0A2W1BD53 A0A0N1I6T9 H9JX83 A0A0N1IBS6 A0A212FIL5 A0A2H1VX30 A0A194R3H3 A0A194PPG4 A0A1Q3FR87 A0A2H1V873 A0A1B6E1F2 A0A1B6D5W8 A0A0T6B9C6 B0WNN7 A0A2A4JEG9 A0A3S2PE71 Q5TQY8 A0A182U0B4 A0A1Y9IRY8 D0VYS1 A0A1Y9GKT0 A0A1Y9J100 G9F9I3 A0A2H1WZM4 D5KQK3 H9JX80 A0A1Y1K2R8 H9IZH5 A0A1S4FLR9 A0A182GJ50 A0A212F963 A0A212EKQ3 D0VYR5 A0A194PMG8 D0VYR6 G9F9G9 D0VYR8 Q16WF2 A0A182QI13 D0VYS0 A0A1S4G3W5 A0A2M3ZET6 A0A194PVV9 A0A1Y9IW73 A0A2M4BKN2 Q16ET6 W5JN46 H9JXC4 A0A212EXF2 A0A212FF93 A0A194R3H6 A0A212FF90 A0A2J7PTJ0 A0A182HAK7 Q8T0W7 S5SP57 A0A2W1BNB0 A0A182J417 A0A182LL23 D2X5V7 A0A3S2TR57 A0A194R360 A0A194QDS5 A0A0N1PFL9 D0VYR9 H9JLY4 A0A194PMT2 A0A2H1VWC6 G9F9I2 D9ILU1 A0A1B0CB38 D6X3T6 A0A1Y1KBB7 A0A182YP84 A0A212FBG8 H9JLY5 A0A2J7PKH6 A0A165G9G6
A0A2A4IWH6 A0A194QH27 A0A194QFH9 H9JXC7 A0A3G1T1E9 A0A194QFC2 H9JX79 H9JXC5 A0A3S2N597 A0A2W1BEU6 A0A2A4IZ96 G9F9G6 A0A2H1WLJ0 A0A2H1WLB6 A0A194QLC3 A0A2W1BJB1 A0A2W1BD53 A0A0N1I6T9 H9JX83 A0A0N1IBS6 A0A212FIL5 A0A2H1VX30 A0A194R3H3 A0A194PPG4 A0A1Q3FR87 A0A2H1V873 A0A1B6E1F2 A0A1B6D5W8 A0A0T6B9C6 B0WNN7 A0A2A4JEG9 A0A3S2PE71 Q5TQY8 A0A182U0B4 A0A1Y9IRY8 D0VYS1 A0A1Y9GKT0 A0A1Y9J100 G9F9I3 A0A2H1WZM4 D5KQK3 H9JX80 A0A1Y1K2R8 H9IZH5 A0A1S4FLR9 A0A182GJ50 A0A212F963 A0A212EKQ3 D0VYR5 A0A194PMG8 D0VYR6 G9F9G9 D0VYR8 Q16WF2 A0A182QI13 D0VYS0 A0A1S4G3W5 A0A2M3ZET6 A0A194PVV9 A0A1Y9IW73 A0A2M4BKN2 Q16ET6 W5JN46 H9JXC4 A0A212EXF2 A0A212FF93 A0A194R3H6 A0A212FF90 A0A2J7PTJ0 A0A182HAK7 Q8T0W7 S5SP57 A0A2W1BNB0 A0A182J417 A0A182LL23 D2X5V7 A0A3S2TR57 A0A194R360 A0A194QDS5 A0A0N1PFL9 D0VYR9 H9JLY4 A0A194PMT2 A0A2H1VWC6 G9F9I2 D9ILU1 A0A1B0CB38 D6X3T6 A0A1Y1KBB7 A0A182YP84 A0A212FBG8 H9JLY5 A0A2J7PKH6 A0A165G9G6
PDB
3VII
E-value=1.20238e-92,
Score=867
Ontologies
GO
PANTHER
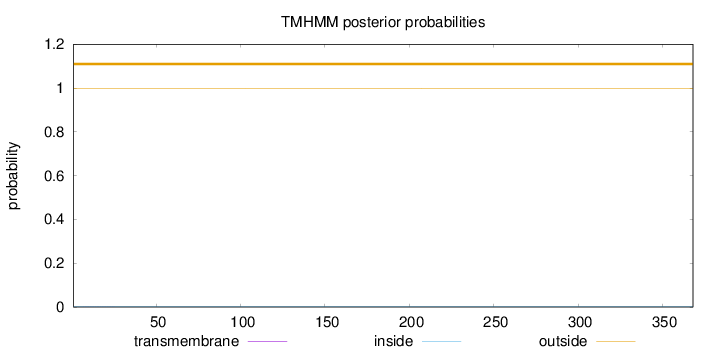
Topology
Length:
368
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00772999999999999
Exp number, first 60 AAs:
0.00414
Total prob of N-in:
0.00237
outside
1 - 368
Population Genetic Test Statistics
Pi
200.521211
Theta
131.873578
Tajima's D
1.332427
CLR
0.436804
CSRT
0.741412929353532
Interpretation
Uncertain
