Gene
KWMTBOMO12230 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014190
Annotation
PREDICTED:_myrosinase_1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.088
Sequence
CDS
ATGACGGTACAAGCAACGGTTGTTTTATGCTTGCTGACGAAGGCGGCTTGGGGCGAGATGAAATTTCCACCTGGCTTCAGATTCGGGGCTGCAACCGCTGCTTATCAGATCGAAGGCTCCTGGAACGTGAGTGATAAAGCCGAGAGCGTATGGGATAGATTTACACATGAACATAAATATTACGTGGACTCTGGAAGCAATGGTGATGTCGCCTGCGACTCCTACAATAAGTGGAAGGATGATGTACGAATTGCAAAGGAGTTGAACTTACACTTCTATAGATTTTCGATCTCGTGGCCTCGTCTACTACCGACTGCGTTTTCAAATAAAATCAGCGACGACGGGAGAAATTACTACAATCAGTTGATCGATGCTCTCCTGGAAGAGGGTATAGAGCCATCGGTGACTCTGTTTCAATTGGATTTGCCTCAAAAATTGCAGGATCTGGGCGGCTGGGCAAACCCTTTAATAATCGATTGGTTCGCCAACTACGCGAGGGTGGTATTTTCTCTTTATGGAGACCGAGTGAAGACCTGGATAACTATTAATGAGCCCCTACTAATTTGTGAGGTGTCGTACAGCGATAGTAAAATGGCCCCTGGAATCAAATCCGTTGACTTAGGAAAATATTTGTGTGCCAAGAATGTTTTGTTAGCTCATGCCACAGCTTGGAGGATTTATGATGAGGAGTTCAGACCGAAGTATCACGGTAAAGTTTCCTTGACGAACGTTTTGATATGGTATGAACCAACTACGGACAACGATCGTGACCTTGCTGACTTGGCGAACCAGTTGTGTAATGGAATATATACGTATCCGATTTTCTCGAAAACCGGTAACTGGCCACAGTCTGTTATAAAAATAGTGGACGAAAGAAGCAAAATGTTGGGATACCCGTACTCTCTTCTCCCTGAGTTTACTAAAGAAGAAATCGAATTTATAAAAGGTACATACGACTTCTTCGGAATGAATTACTACATTTCAAAGTCAATAAGGAAGCCTCTGGTTAACGAACATATACCGTTGTGGCCTGTGGGAGTACCGATATTGGGTGCCATTTTGGAGAGTCCACCCACTTGGACGAATGGAGCTACGAAGTGGTTTTCAGTAAAATACATTTGTTCTTACGTAGCTTAA
Protein
MTVQATVVLCLLTKAAWGEMKFPPGFRFGAATAAYQIEGSWNVSDKAESVWDRFTHEHKYYVDSGSNGDVACDSYNKWKDDVRIAKELNLHFYRFSISWPRLLPTAFSNKISDDGRNYYNQLIDALLEEGIEPSVTLFQLDLPQKLQDLGGWANPLIIDWFANYARVVFSLYGDRVKTWITINEPLLICEVSYSDSKMAPGIKSVDLGKYLCAKNVLLAHATAWRIYDEEFRPKYHGKVSLTNVLIWYEPTTDNDRDLADLANQLCNGIYTYPIFSKTGNWPQSVIKIVDERSKMLGYPYSLLPEFTKEEIEFIKGTYDFFGMNYYISKSIRKPLVNEHIPLWPVGVPILGAILESPPTWTNGATKWFSVKYICSYVA
Summary
Similarity
Belongs to the glycosyl hydrolase 1 family.
Belongs to the class-I aminoacyl-tRNA synthetase family.
Belongs to the class-I aminoacyl-tRNA synthetase family.
Uniprot
H9JXC5
H9JX79
A0A194QLC3
A0A194QH27
A0A2W1BMB0
H9JXC6
+ More
A0A0N1IBS6 A0A3G1T1E9 A0A2A4J905 A0A2W1BC39 A0A2A4IWH6 A0A3S2N597 A0A2H1VC49 A0A194QFH9 A0A2A4IZ96 A0A194QFC2 H9JXC4 A0A2H1WZM4 A0A2W1BJB1 A0A2H1WLB6 A0A2W1BEU6 A0A2H1W6N8 H9JXC2 A0A0N1I6T9 H9JX80 A0A2H1WLJ0 G9F9G9 A0A194R3H3 A0A194PPG4 A0A3S2PE71 A0A194R3H6 H9JXC7 G9F9I3 A0A2H1VX30 A0A194PMG8 A0A2A4JPS5 A0A212FF90 A0A212FF93 A0A194PMT8 A0A212FBG8 A0A3S2NZ50 A0A1Y9GKT0 Q5TQY8 A0A194PNS3 A0A2A4JPI4 A0A2W1BWF3 A0A2A4IYR0 A0A194R8P7 A0A212FKW0 A0A1Y1K666 A0A194PPG9 H9JLY4 A0A2A4IZI1 B0WNN7 A0A194RQF8 A0A1Y1KBI8 A0A194R4S7 A0A2H1VF65 D0VYR9 A0A182U0B4 A0A1Y9IRY8 A0A194R365 A0A194PMT2 A0A194PJU3 A0A194R400 A0A1Y1K6D9 G9F9G4 A0A212EPH6 A0A212FIL5 A0A2H1V873 D0VYS0 A0A195EE04 A0A182T2U8 A0A2A4JPN8 H9JLY5 A0A1Y9IW73 D0VYS1 A0A1L8DQN1 A0A194R360 A0A182YP84 A0A2W1BD53 D0VYR8 D6X3T6 H9JMQ9 D6WPV3 J3JYA4 A0A194PRE4 A0A1S4G3W5 A0A212F3B1 A0A2A4JEG9 A0A026WLX1 A0A034VAL4 A0A194PU18 A0A182JGM2 D0VYR6 Q16WF2 A0A182LL23 D0VYR5 A0A034VEY7
A0A0N1IBS6 A0A3G1T1E9 A0A2A4J905 A0A2W1BC39 A0A2A4IWH6 A0A3S2N597 A0A2H1VC49 A0A194QFH9 A0A2A4IZ96 A0A194QFC2 H9JXC4 A0A2H1WZM4 A0A2W1BJB1 A0A2H1WLB6 A0A2W1BEU6 A0A2H1W6N8 H9JXC2 A0A0N1I6T9 H9JX80 A0A2H1WLJ0 G9F9G9 A0A194R3H3 A0A194PPG4 A0A3S2PE71 A0A194R3H6 H9JXC7 G9F9I3 A0A2H1VX30 A0A194PMG8 A0A2A4JPS5 A0A212FF90 A0A212FF93 A0A194PMT8 A0A212FBG8 A0A3S2NZ50 A0A1Y9GKT0 Q5TQY8 A0A194PNS3 A0A2A4JPI4 A0A2W1BWF3 A0A2A4IYR0 A0A194R8P7 A0A212FKW0 A0A1Y1K666 A0A194PPG9 H9JLY4 A0A2A4IZI1 B0WNN7 A0A194RQF8 A0A1Y1KBI8 A0A194R4S7 A0A2H1VF65 D0VYR9 A0A182U0B4 A0A1Y9IRY8 A0A194R365 A0A194PMT2 A0A194PJU3 A0A194R400 A0A1Y1K6D9 G9F9G4 A0A212EPH6 A0A212FIL5 A0A2H1V873 D0VYS0 A0A195EE04 A0A182T2U8 A0A2A4JPN8 H9JLY5 A0A1Y9IW73 D0VYS1 A0A1L8DQN1 A0A194R360 A0A182YP84 A0A2W1BD53 D0VYR8 D6X3T6 H9JMQ9 D6WPV3 J3JYA4 A0A194PRE4 A0A1S4G3W5 A0A212F3B1 A0A2A4JEG9 A0A026WLX1 A0A034VAL4 A0A194PU18 A0A182JGM2 D0VYR6 Q16WF2 A0A182LL23 D0VYR5 A0A034VEY7
Pubmed
EMBL
BABH01035826
BABH01035827
BABH01035825
KQ459053
KPJ04241.1
KPJ04240.1
+ More
KZ150152 PZC72823.1 BABH01035833 BABH01035834 BABH01035835 KQ461157 KPJ08092.1 MG992371 AXY94809.1 NWSH01002416 PCG68339.1 PZC72822.1 NWSH01005512 PCG64155.1 RSAL01000405 RVE41892.1 ODYU01001752 SOQ38385.1 KPJ04242.1 NWSH01004417 PCG65145.1 KPJ04243.1 BABH01035821 ODYU01012291 SOQ58525.1 PZC72820.1 ODYU01009444 SOQ53875.1 PZC72831.1 ODYU01006661 SOQ48703.1 BABH01035819 BABH01035820 KPJ08090.1 BABH01035816 BABH01035817 SOQ53877.1 JN033714 AEW46867.1 KQ460779 KPJ12242.1 KQ459598 KPI94624.1 RSAL01000077 RVE48784.1 KPJ12247.1 BABH01035838 BABH01035839 JN033728 AEW46881.1 ODYU01004975 SOQ45390.1 KPI94631.1 NWSH01000819 PCG74065.1 AGBW02008847 OWR52398.1 OWR52397.1 KPI94632.1 AGBW02009344 OWR51063.1 RSAL01000019 RVE52797.1 APCN01002262 AAAB01008960 EAL40075.2 KPI94633.1 NWSH01000883 PCG73729.1 KZ149936 PZC77130.1 NWSH01004958 PCG64548.1 KPJ12246.1 AGBW02007954 OWR54377.1 GEZM01091301 JAV56942.1 KPI94629.1 BABH01005437 NWSH01004583 PCG64896.1 DS232013 EDS31873.1 KQ459875 KPJ19634.1 GEZM01091305 JAV56935.1 KPJ12245.1 ODYU01001971 SOQ38904.1 AB508958 BAI50022.1 KPJ12248.1 KPI94627.1 KQ459601 KPI93701.1 KPJ12249.1 GEZM01091306 JAV56934.1 JN033709 AEW46862.1 AGBW02013459 OWR43376.1 AGBW02008370 OWR53561.1 ODYU01001184 SOQ37050.1 AB508959 BAI50023.1 KQ979074 KYN23032.1 PCG73726.1 BABH01005438 AB508960 BAI50024.1 GFDF01005306 JAV08778.1 KPJ12243.1 KZ150122 PZC73162.1 AB508957 BAI50021.1 KQ971326 EEZ97373.2 BABH01016738 KQ971354 EFA06168.1 APGK01045791 APGK01045792 APGK01045793 BT128228 KB741039 AEE63189.1 ENN74525.1 KPI93700.1 AGBW02010585 OWR48225.1 NWSH01001829 PCG69984.1 KK107159 EZA56631.1 GAKP01018616 JAC40336.1 KPI94630.1 AB508955 BAI50019.1 CH477568 EAT38910.1 AB508954 AB508956 BAI50018.1 BAI50020.1 GAKP01018617 JAC40335.1
KZ150152 PZC72823.1 BABH01035833 BABH01035834 BABH01035835 KQ461157 KPJ08092.1 MG992371 AXY94809.1 NWSH01002416 PCG68339.1 PZC72822.1 NWSH01005512 PCG64155.1 RSAL01000405 RVE41892.1 ODYU01001752 SOQ38385.1 KPJ04242.1 NWSH01004417 PCG65145.1 KPJ04243.1 BABH01035821 ODYU01012291 SOQ58525.1 PZC72820.1 ODYU01009444 SOQ53875.1 PZC72831.1 ODYU01006661 SOQ48703.1 BABH01035819 BABH01035820 KPJ08090.1 BABH01035816 BABH01035817 SOQ53877.1 JN033714 AEW46867.1 KQ460779 KPJ12242.1 KQ459598 KPI94624.1 RSAL01000077 RVE48784.1 KPJ12247.1 BABH01035838 BABH01035839 JN033728 AEW46881.1 ODYU01004975 SOQ45390.1 KPI94631.1 NWSH01000819 PCG74065.1 AGBW02008847 OWR52398.1 OWR52397.1 KPI94632.1 AGBW02009344 OWR51063.1 RSAL01000019 RVE52797.1 APCN01002262 AAAB01008960 EAL40075.2 KPI94633.1 NWSH01000883 PCG73729.1 KZ149936 PZC77130.1 NWSH01004958 PCG64548.1 KPJ12246.1 AGBW02007954 OWR54377.1 GEZM01091301 JAV56942.1 KPI94629.1 BABH01005437 NWSH01004583 PCG64896.1 DS232013 EDS31873.1 KQ459875 KPJ19634.1 GEZM01091305 JAV56935.1 KPJ12245.1 ODYU01001971 SOQ38904.1 AB508958 BAI50022.1 KPJ12248.1 KPI94627.1 KQ459601 KPI93701.1 KPJ12249.1 GEZM01091306 JAV56934.1 JN033709 AEW46862.1 AGBW02013459 OWR43376.1 AGBW02008370 OWR53561.1 ODYU01001184 SOQ37050.1 AB508959 BAI50023.1 KQ979074 KYN23032.1 PCG73726.1 BABH01005438 AB508960 BAI50024.1 GFDF01005306 JAV08778.1 KPJ12243.1 KZ150122 PZC73162.1 AB508957 BAI50021.1 KQ971326 EEZ97373.2 BABH01016738 KQ971354 EFA06168.1 APGK01045791 APGK01045792 APGK01045793 BT128228 KB741039 AEE63189.1 ENN74525.1 KPI93700.1 AGBW02010585 OWR48225.1 NWSH01001829 PCG69984.1 KK107159 EZA56631.1 GAKP01018616 JAC40336.1 KPI94630.1 AB508955 BAI50019.1 CH477568 EAT38910.1 AB508954 AB508956 BAI50018.1 BAI50020.1 GAKP01018617 JAC40335.1
Proteomes
Pfam
Interpro
IPR033132
Glyco_hydro_1_N_CS
+ More
IPR001360 Glyco_hydro_1
IPR017853 Glycoside_hydrolase_SF
IPR018120 Glyco_hydro_1_AS
IPR000873 AMP-dep_Synth/Lig
IPR025110 AMP-bd_C
IPR002300 aa-tRNA-synth_Ia
IPR014729 Rossmann-like_a/b/a_fold
IPR001412 aa-tRNA-synth_I_CS
IPR009008 Val/Leu/Ile-tRNA-synth_edit
IPR002301 Ile-tRNA-ligase
IPR033708 Anticodon_Ile_BEm
IPR009080 tRNAsynth_Ia_anticodon-bd
IPR013155 M/V/L/I-tRNA-synth_anticd-bd
IPR001360 Glyco_hydro_1
IPR017853 Glycoside_hydrolase_SF
IPR018120 Glyco_hydro_1_AS
IPR000873 AMP-dep_Synth/Lig
IPR025110 AMP-bd_C
IPR002300 aa-tRNA-synth_Ia
IPR014729 Rossmann-like_a/b/a_fold
IPR001412 aa-tRNA-synth_I_CS
IPR009008 Val/Leu/Ile-tRNA-synth_edit
IPR002301 Ile-tRNA-ligase
IPR033708 Anticodon_Ile_BEm
IPR009080 tRNAsynth_Ia_anticodon-bd
IPR013155 M/V/L/I-tRNA-synth_anticd-bd
Gene 3D
ProteinModelPortal
H9JXC5
H9JX79
A0A194QLC3
A0A194QH27
A0A2W1BMB0
H9JXC6
+ More
A0A0N1IBS6 A0A3G1T1E9 A0A2A4J905 A0A2W1BC39 A0A2A4IWH6 A0A3S2N597 A0A2H1VC49 A0A194QFH9 A0A2A4IZ96 A0A194QFC2 H9JXC4 A0A2H1WZM4 A0A2W1BJB1 A0A2H1WLB6 A0A2W1BEU6 A0A2H1W6N8 H9JXC2 A0A0N1I6T9 H9JX80 A0A2H1WLJ0 G9F9G9 A0A194R3H3 A0A194PPG4 A0A3S2PE71 A0A194R3H6 H9JXC7 G9F9I3 A0A2H1VX30 A0A194PMG8 A0A2A4JPS5 A0A212FF90 A0A212FF93 A0A194PMT8 A0A212FBG8 A0A3S2NZ50 A0A1Y9GKT0 Q5TQY8 A0A194PNS3 A0A2A4JPI4 A0A2W1BWF3 A0A2A4IYR0 A0A194R8P7 A0A212FKW0 A0A1Y1K666 A0A194PPG9 H9JLY4 A0A2A4IZI1 B0WNN7 A0A194RQF8 A0A1Y1KBI8 A0A194R4S7 A0A2H1VF65 D0VYR9 A0A182U0B4 A0A1Y9IRY8 A0A194R365 A0A194PMT2 A0A194PJU3 A0A194R400 A0A1Y1K6D9 G9F9G4 A0A212EPH6 A0A212FIL5 A0A2H1V873 D0VYS0 A0A195EE04 A0A182T2U8 A0A2A4JPN8 H9JLY5 A0A1Y9IW73 D0VYS1 A0A1L8DQN1 A0A194R360 A0A182YP84 A0A2W1BD53 D0VYR8 D6X3T6 H9JMQ9 D6WPV3 J3JYA4 A0A194PRE4 A0A1S4G3W5 A0A212F3B1 A0A2A4JEG9 A0A026WLX1 A0A034VAL4 A0A194PU18 A0A182JGM2 D0VYR6 Q16WF2 A0A182LL23 D0VYR5 A0A034VEY7
A0A0N1IBS6 A0A3G1T1E9 A0A2A4J905 A0A2W1BC39 A0A2A4IWH6 A0A3S2N597 A0A2H1VC49 A0A194QFH9 A0A2A4IZ96 A0A194QFC2 H9JXC4 A0A2H1WZM4 A0A2W1BJB1 A0A2H1WLB6 A0A2W1BEU6 A0A2H1W6N8 H9JXC2 A0A0N1I6T9 H9JX80 A0A2H1WLJ0 G9F9G9 A0A194R3H3 A0A194PPG4 A0A3S2PE71 A0A194R3H6 H9JXC7 G9F9I3 A0A2H1VX30 A0A194PMG8 A0A2A4JPS5 A0A212FF90 A0A212FF93 A0A194PMT8 A0A212FBG8 A0A3S2NZ50 A0A1Y9GKT0 Q5TQY8 A0A194PNS3 A0A2A4JPI4 A0A2W1BWF3 A0A2A4IYR0 A0A194R8P7 A0A212FKW0 A0A1Y1K666 A0A194PPG9 H9JLY4 A0A2A4IZI1 B0WNN7 A0A194RQF8 A0A1Y1KBI8 A0A194R4S7 A0A2H1VF65 D0VYR9 A0A182U0B4 A0A1Y9IRY8 A0A194R365 A0A194PMT2 A0A194PJU3 A0A194R400 A0A1Y1K6D9 G9F9G4 A0A212EPH6 A0A212FIL5 A0A2H1V873 D0VYS0 A0A195EE04 A0A182T2U8 A0A2A4JPN8 H9JLY5 A0A1Y9IW73 D0VYS1 A0A1L8DQN1 A0A194R360 A0A182YP84 A0A2W1BD53 D0VYR8 D6X3T6 H9JMQ9 D6WPV3 J3JYA4 A0A194PRE4 A0A1S4G3W5 A0A212F3B1 A0A2A4JEG9 A0A026WLX1 A0A034VAL4 A0A194PU18 A0A182JGM2 D0VYR6 Q16WF2 A0A182LL23 D0VYR5 A0A034VEY7
PDB
5CG0
E-value=3.79869e-83,
Score=785
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.916094
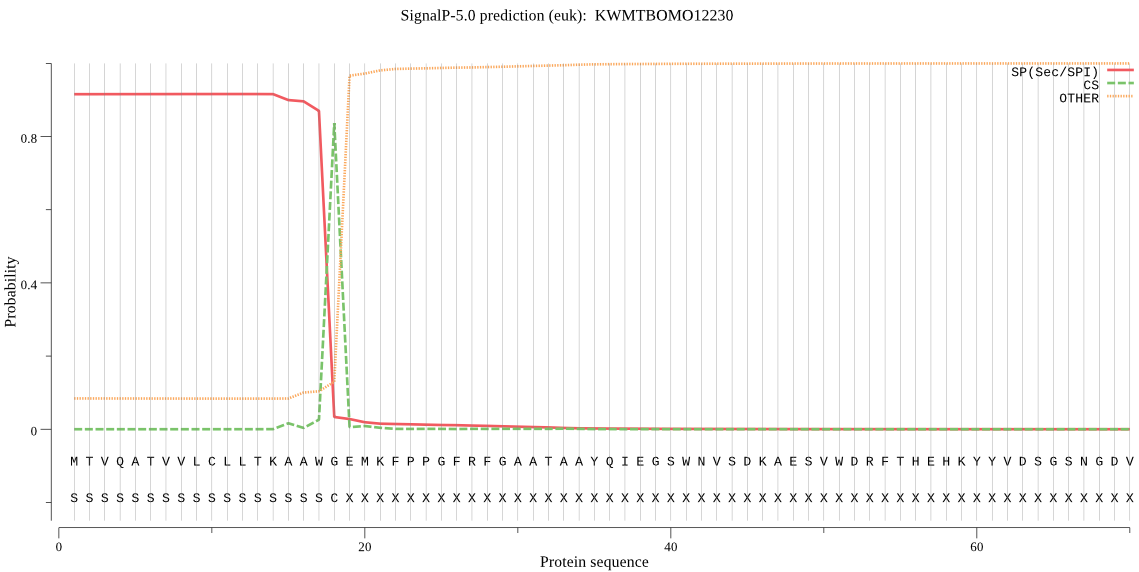
Length:
378
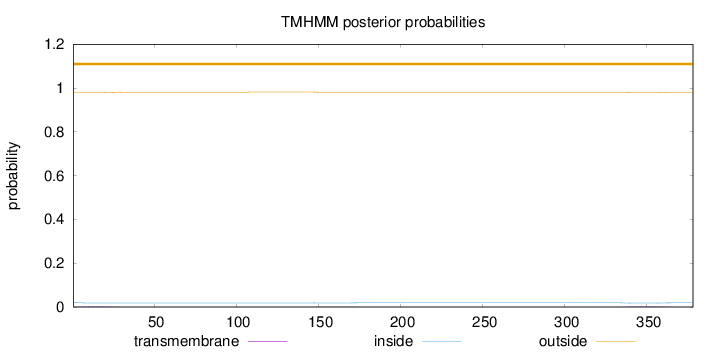
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15098
Exp number, first 60 AAs:
0.05824
Total prob of N-in:
0.02168
outside
1 - 378
Population Genetic Test Statistics
Pi
251.236267
Theta
154.555248
Tajima's D
2.820064
CLR
0.292617
CSRT
0.969851507424629
Interpretation
Uncertain
