Pre Gene Modal
BGIBMGA014144
Annotation
PREDICTED:_myrosinase_1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.559
Sequence
CDS
ATGTCGTACAGCGATAGTAATCTGGCCCCTGGAATCGAATCCATTGAATTAGGAAATTATTTGTGTGCCAAGAATGTTTTGTTAGCTCATGCCACAGCTTGGAGGATTTATGATGAGGAGTTCAGACCGAAGTATCACGGTAAAGTTTCCTTGACGAACATTTTGATATGGTATGAACCAACTACGGGCAACGATAGTAACCTTGGTAACTTGGCGAACCAGTTGTGTAATGGAATATATACGCATCCGATTTTCTCGAAAACTGGTAACTGGCCACAGTCTGTTATAAAAATAGTGGACGAAAGAAGCAAATTGTTGGGATATCCTTACTCTCTTCTTCCTGAGTTTACTAAAAAAGAAATAGAATTTATAAAAGGTACTTACGACTTCTTCGGAATGAATTACTACATTTCAAAGTCAATAAGGAAGTCCCTGGTAAATGAACATATACCGTTGTGGCCTCTTGAGGGAGTACCGATATTGGGAGCCATTTTGGAAAGTCAACCCACTTGGACGAGCGGAGCTACGAAGTGGTTTTCGGTAAAATACATTTGTTTTTACGTACCTTAA
Protein
MSYSDSNLAPGIESIELGNYLCAKNVLLAHATAWRIYDEEFRPKYHGKVSLTNILIWYEPTTGNDSNLGNLANQLCNGIYTHPIFSKTGNWPQSVIKIVDERSKLLGYPYSLLPEFTKKEIEFIKGTYDFFGMNYYISKSIRKSLVNEHIPLWPLEGVPILGAILESQPTWTSGATKWFSVKYICFYVP
Summary
Similarity
Belongs to the glycosyl hydrolase 1 family.
Belongs to the class-I aminoacyl-tRNA synthetase family.
Belongs to the class-I aminoacyl-tRNA synthetase family.
Uniprot
H9JX79
H9JXC5
A0A194QLC3
A0A2H1VC49
G9F9G6
H9JXC7
+ More
A0A194QH27 A0A2A4J905 A0A2W1BMB0 A0A0N1PFL9 A0A2A4IWH6 A0A2W1BC39 H9JXC6 A0A0N1IBS6 A0A194QFH9 A0A3G1T1E9 A0A2A4IZ96 H9JXC2 A0A3S2N597 A0A2W1BEU6 A0A2H1W6N8 A0A2H1WLJ0 A0A194QFC2 A0A2W1BJB1 H9JXC4 A0A2H1WZM4 A0A2H1WLB6 A0A2H1VX30 V5GUF1 V5GII4 V5GSR2 A0A212FBG8 A0A212FIL5 A0A0K8TUU4 D0VYR9 A0A3S2NZ50 D0VYS0 A0A194R3H6 A0A194PPG4 A0A0B5EM11 H9JX80 D0VYR8 H9JX83 A0A194R3H3 D0VYS1 D0VYR6 A0A194PMG8 G9F9G9 A0A0B5EKM7 A0A194QDS5 D0VYR5 A0A059U9B2 D2X5V7 S5SP57 A0A194R3Z4 A0A194R8P7 B0WNN7 A0A1Q3FR87 X2CIK2 A0A059UAE7 D6WPV3 A0A3S2PE71 G9F9I3 A0A212FF90 A0A060NZS1 A0A212FF93 V5I949 A0A1W4WG49 A0A0S7EMZ9 N6TKX9 J3JV61 V5I873 A0A2A4JPN8 A0A0N1I6T9 A0A1L8DQN1 D9ILU1 A0A1L8DQS5 A0A194PU18 A0A1L8DR18 A0A2A4JEG9 A0A2M4BKN2 A0A2H1V873 A0A212EXF2 A0A2H1VVL0 A0A194PMT8 A0A1S4FLR9 A0A2M3ZET6 A0A059U5M3 S4NTM4 A0A2A4JPS5 A0A1B0CB38 V5GH04 A0A1W4WMX7 A0A2W1BD53 W5JN46 A0A165G9G6 A0A1B0DHF5
A0A194QH27 A0A2A4J905 A0A2W1BMB0 A0A0N1PFL9 A0A2A4IWH6 A0A2W1BC39 H9JXC6 A0A0N1IBS6 A0A194QFH9 A0A3G1T1E9 A0A2A4IZ96 H9JXC2 A0A3S2N597 A0A2W1BEU6 A0A2H1W6N8 A0A2H1WLJ0 A0A194QFC2 A0A2W1BJB1 H9JXC4 A0A2H1WZM4 A0A2H1WLB6 A0A2H1VX30 V5GUF1 V5GII4 V5GSR2 A0A212FBG8 A0A212FIL5 A0A0K8TUU4 D0VYR9 A0A3S2NZ50 D0VYS0 A0A194R3H6 A0A194PPG4 A0A0B5EM11 H9JX80 D0VYR8 H9JX83 A0A194R3H3 D0VYS1 D0VYR6 A0A194PMG8 G9F9G9 A0A0B5EKM7 A0A194QDS5 D0VYR5 A0A059U9B2 D2X5V7 S5SP57 A0A194R3Z4 A0A194R8P7 B0WNN7 A0A1Q3FR87 X2CIK2 A0A059UAE7 D6WPV3 A0A3S2PE71 G9F9I3 A0A212FF90 A0A060NZS1 A0A212FF93 V5I949 A0A1W4WG49 A0A0S7EMZ9 N6TKX9 J3JV61 V5I873 A0A2A4JPN8 A0A0N1I6T9 A0A1L8DQN1 D9ILU1 A0A1L8DQS5 A0A194PU18 A0A1L8DR18 A0A2A4JEG9 A0A2M4BKN2 A0A2H1V873 A0A212EXF2 A0A2H1VVL0 A0A194PMT8 A0A1S4FLR9 A0A2M3ZET6 A0A059U5M3 S4NTM4 A0A2A4JPS5 A0A1B0CB38 V5GH04 A0A1W4WMX7 A0A2W1BD53 W5JN46 A0A165G9G6 A0A1B0DHF5
Pubmed
EMBL
BABH01035825
BABH01035826
BABH01035827
KQ459053
KPJ04241.1
ODYU01001752
+ More
SOQ38385.1 JN033711 AEW46864.1 BABH01035838 BABH01035839 KPJ04240.1 NWSH01002416 PCG68339.1 KZ150152 PZC72823.1 KQ461157 KPJ08091.1 NWSH01005512 PCG64155.1 PZC72822.1 BABH01035833 BABH01035834 BABH01035835 KPJ08092.1 KPJ04242.1 MG992371 AXY94809.1 NWSH01004417 PCG65145.1 BABH01035819 BABH01035820 RSAL01000405 RVE41892.1 PZC72831.1 ODYU01006661 SOQ48703.1 ODYU01009444 SOQ53877.1 KPJ04243.1 PZC72820.1 BABH01035821 ODYU01012291 SOQ58525.1 SOQ53875.1 ODYU01004975 SOQ45390.1 GALX01004593 JAB63873.1 GALX01004592 JAB63874.1 GALX01005183 JAB63283.1 AGBW02009344 OWR51063.1 AGBW02008370 OWR53561.1 GBUI01000003 JAI18232.1 AB508958 BAI50022.1 RSAL01000019 RVE52797.1 AB508959 BAI50023.1 KQ460779 KPJ12247.1 KQ459598 KPI94624.1 KP068699 AJE75670.1 BABH01035816 BABH01035817 AB508957 BAI50021.1 BABH01035811 BABH01035812 KPJ12242.1 AB508960 BAI50024.1 AB508955 BAI50019.1 KPI94631.1 JN033714 AEW46867.1 KP068693 AJE75664.1 KQ459144 KPJ03612.1 AB508954 AB508956 BAI50018.1 BAI50020.1 KF377835 AHZ59653.1 GQ911585 ADB23476.1 KC891004 AGS32242.1 KPJ12244.1 KPJ12246.1 DS232013 EDS31873.1 GFDL01004884 JAV30161.1 KC533772 AGP76180.1 KF377848 AHZ59666.1 KQ971354 EFA06168.1 RSAL01000077 RVE48784.1 JN033728 AEW46881.1 AGBW02008847 OWR52398.1 AB915865 BAO85043.1 OWR52397.1 GALX01003555 JAB64911.1 GBUH01000007 JAO03635.1 APGK01033064 APGK01033065 APGK01033066 KB740860 ENN78593.1 BT127127 AEE62089.1 GALX01005185 JAB63281.1 NWSH01000883 PCG73726.1 KPJ08090.1 GFDF01005306 JAV08778.1 HM152540 ADK12988.1 GFDF01005305 JAV08779.1 KPI94630.1 GFDF01005304 JAV08780.1 NWSH01001829 PCG69984.1 GGFJ01004382 MBW53523.1 ODYU01001184 SOQ37050.1 AGBW02011758 OWR46178.1 ODYU01004710 SOQ44881.1 KPI94632.1 GGFM01006254 MBW27005.1 KF377834 AHZ59652.1 GAIX01010404 JAA82156.1 NWSH01000819 PCG74065.1 AJWK01004788 AJWK01004789 AJWK01004790 GALX01005182 JAB63284.1 KZ150122 PZC73162.1 ADMH02000892 ETN64738.1 KU886065 AMY26906.1 AJVK01061043
SOQ38385.1 JN033711 AEW46864.1 BABH01035838 BABH01035839 KPJ04240.1 NWSH01002416 PCG68339.1 KZ150152 PZC72823.1 KQ461157 KPJ08091.1 NWSH01005512 PCG64155.1 PZC72822.1 BABH01035833 BABH01035834 BABH01035835 KPJ08092.1 KPJ04242.1 MG992371 AXY94809.1 NWSH01004417 PCG65145.1 BABH01035819 BABH01035820 RSAL01000405 RVE41892.1 PZC72831.1 ODYU01006661 SOQ48703.1 ODYU01009444 SOQ53877.1 KPJ04243.1 PZC72820.1 BABH01035821 ODYU01012291 SOQ58525.1 SOQ53875.1 ODYU01004975 SOQ45390.1 GALX01004593 JAB63873.1 GALX01004592 JAB63874.1 GALX01005183 JAB63283.1 AGBW02009344 OWR51063.1 AGBW02008370 OWR53561.1 GBUI01000003 JAI18232.1 AB508958 BAI50022.1 RSAL01000019 RVE52797.1 AB508959 BAI50023.1 KQ460779 KPJ12247.1 KQ459598 KPI94624.1 KP068699 AJE75670.1 BABH01035816 BABH01035817 AB508957 BAI50021.1 BABH01035811 BABH01035812 KPJ12242.1 AB508960 BAI50024.1 AB508955 BAI50019.1 KPI94631.1 JN033714 AEW46867.1 KP068693 AJE75664.1 KQ459144 KPJ03612.1 AB508954 AB508956 BAI50018.1 BAI50020.1 KF377835 AHZ59653.1 GQ911585 ADB23476.1 KC891004 AGS32242.1 KPJ12244.1 KPJ12246.1 DS232013 EDS31873.1 GFDL01004884 JAV30161.1 KC533772 AGP76180.1 KF377848 AHZ59666.1 KQ971354 EFA06168.1 RSAL01000077 RVE48784.1 JN033728 AEW46881.1 AGBW02008847 OWR52398.1 AB915865 BAO85043.1 OWR52397.1 GALX01003555 JAB64911.1 GBUH01000007 JAO03635.1 APGK01033064 APGK01033065 APGK01033066 KB740860 ENN78593.1 BT127127 AEE62089.1 GALX01005185 JAB63281.1 NWSH01000883 PCG73726.1 KPJ08090.1 GFDF01005306 JAV08778.1 HM152540 ADK12988.1 GFDF01005305 JAV08779.1 KPI94630.1 GFDF01005304 JAV08780.1 NWSH01001829 PCG69984.1 GGFJ01004382 MBW53523.1 ODYU01001184 SOQ37050.1 AGBW02011758 OWR46178.1 ODYU01004710 SOQ44881.1 KPI94632.1 GGFM01006254 MBW27005.1 KF377834 AHZ59652.1 GAIX01010404 JAA82156.1 NWSH01000819 PCG74065.1 AJWK01004788 AJWK01004789 AJWK01004790 GALX01005182 JAB63284.1 KZ150122 PZC73162.1 ADMH02000892 ETN64738.1 KU886065 AMY26906.1 AJVK01061043
Proteomes
Pfam
Interpro
IPR017853
Glycoside_hydrolase_SF
+ More
IPR001360 Glyco_hydro_1
IPR033132 Glyco_hydro_1_N_CS
IPR018120 Glyco_hydro_1_AS
IPR000873 AMP-dep_Synth/Lig
IPR025110 AMP-bd_C
IPR002300 aa-tRNA-synth_Ia
IPR014729 Rossmann-like_a/b/a_fold
IPR001412 aa-tRNA-synth_I_CS
IPR009008 Val/Leu/Ile-tRNA-synth_edit
IPR002301 Ile-tRNA-ligase
IPR011234 Fumarylacetoacetase-like_C
IPR036663 Fumarylacetoacetase_C_sf
IPR001360 Glyco_hydro_1
IPR033132 Glyco_hydro_1_N_CS
IPR018120 Glyco_hydro_1_AS
IPR000873 AMP-dep_Synth/Lig
IPR025110 AMP-bd_C
IPR002300 aa-tRNA-synth_Ia
IPR014729 Rossmann-like_a/b/a_fold
IPR001412 aa-tRNA-synth_I_CS
IPR009008 Val/Leu/Ile-tRNA-synth_edit
IPR002301 Ile-tRNA-ligase
IPR011234 Fumarylacetoacetase-like_C
IPR036663 Fumarylacetoacetase_C_sf
Gene 3D
ProteinModelPortal
H9JX79
H9JXC5
A0A194QLC3
A0A2H1VC49
G9F9G6
H9JXC7
+ More
A0A194QH27 A0A2A4J905 A0A2W1BMB0 A0A0N1PFL9 A0A2A4IWH6 A0A2W1BC39 H9JXC6 A0A0N1IBS6 A0A194QFH9 A0A3G1T1E9 A0A2A4IZ96 H9JXC2 A0A3S2N597 A0A2W1BEU6 A0A2H1W6N8 A0A2H1WLJ0 A0A194QFC2 A0A2W1BJB1 H9JXC4 A0A2H1WZM4 A0A2H1WLB6 A0A2H1VX30 V5GUF1 V5GII4 V5GSR2 A0A212FBG8 A0A212FIL5 A0A0K8TUU4 D0VYR9 A0A3S2NZ50 D0VYS0 A0A194R3H6 A0A194PPG4 A0A0B5EM11 H9JX80 D0VYR8 H9JX83 A0A194R3H3 D0VYS1 D0VYR6 A0A194PMG8 G9F9G9 A0A0B5EKM7 A0A194QDS5 D0VYR5 A0A059U9B2 D2X5V7 S5SP57 A0A194R3Z4 A0A194R8P7 B0WNN7 A0A1Q3FR87 X2CIK2 A0A059UAE7 D6WPV3 A0A3S2PE71 G9F9I3 A0A212FF90 A0A060NZS1 A0A212FF93 V5I949 A0A1W4WG49 A0A0S7EMZ9 N6TKX9 J3JV61 V5I873 A0A2A4JPN8 A0A0N1I6T9 A0A1L8DQN1 D9ILU1 A0A1L8DQS5 A0A194PU18 A0A1L8DR18 A0A2A4JEG9 A0A2M4BKN2 A0A2H1V873 A0A212EXF2 A0A2H1VVL0 A0A194PMT8 A0A1S4FLR9 A0A2M3ZET6 A0A059U5M3 S4NTM4 A0A2A4JPS5 A0A1B0CB38 V5GH04 A0A1W4WMX7 A0A2W1BD53 W5JN46 A0A165G9G6 A0A1B0DHF5
A0A194QH27 A0A2A4J905 A0A2W1BMB0 A0A0N1PFL9 A0A2A4IWH6 A0A2W1BC39 H9JXC6 A0A0N1IBS6 A0A194QFH9 A0A3G1T1E9 A0A2A4IZ96 H9JXC2 A0A3S2N597 A0A2W1BEU6 A0A2H1W6N8 A0A2H1WLJ0 A0A194QFC2 A0A2W1BJB1 H9JXC4 A0A2H1WZM4 A0A2H1WLB6 A0A2H1VX30 V5GUF1 V5GII4 V5GSR2 A0A212FBG8 A0A212FIL5 A0A0K8TUU4 D0VYR9 A0A3S2NZ50 D0VYS0 A0A194R3H6 A0A194PPG4 A0A0B5EM11 H9JX80 D0VYR8 H9JX83 A0A194R3H3 D0VYS1 D0VYR6 A0A194PMG8 G9F9G9 A0A0B5EKM7 A0A194QDS5 D0VYR5 A0A059U9B2 D2X5V7 S5SP57 A0A194R3Z4 A0A194R8P7 B0WNN7 A0A1Q3FR87 X2CIK2 A0A059UAE7 D6WPV3 A0A3S2PE71 G9F9I3 A0A212FF90 A0A060NZS1 A0A212FF93 V5I949 A0A1W4WG49 A0A0S7EMZ9 N6TKX9 J3JV61 V5I873 A0A2A4JPN8 A0A0N1I6T9 A0A1L8DQN1 D9ILU1 A0A1L8DQS5 A0A194PU18 A0A1L8DR18 A0A2A4JEG9 A0A2M4BKN2 A0A2H1V873 A0A212EXF2 A0A2H1VVL0 A0A194PMT8 A0A1S4FLR9 A0A2M3ZET6 A0A059U5M3 S4NTM4 A0A2A4JPS5 A0A1B0CB38 V5GH04 A0A1W4WMX7 A0A2W1BD53 W5JN46 A0A165G9G6 A0A1B0DHF5
PDB
5CG0
E-value=9.10046e-27,
Score=295
Ontologies
GO
PANTHER
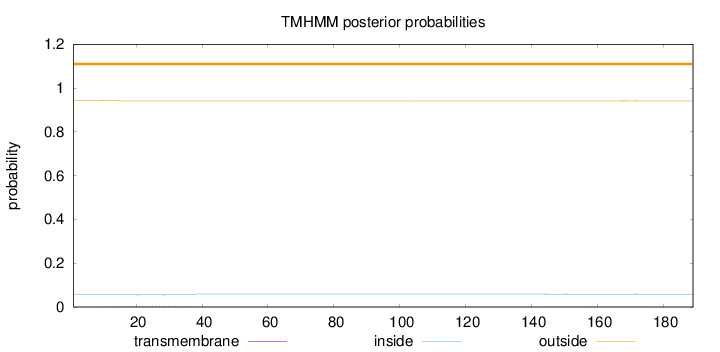
Topology
Length:
189
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0968599999999999
Exp number, first 60 AAs:
0.0825799999999999
Total prob of N-in:
0.05605
outside
1 - 189
Population Genetic Test Statistics
Pi
187.639729
Theta
153.528211
Tajima's D
1.710826
CLR
0.292275
CSRT
0.831508424578771
Interpretation
Uncertain
